Hoelz Lab: Publications
Figure 1. Overview of the NPC. (a) Schematic representation of the NPC architecture. A cutaway view depicting half of an NPC is shown. (b) Comparison of sizes of the NPC with other membrane transporters and example nucleocytoplasmic transport cargoes. Surface representations of human Aquaporin-1 (PDB 1FQY), bacterial SecYEG (PDB 1RHZ), Drosophila melanogaster Importin13–Mago–Y14 (PDB 2X1G), Crm1–Ran(GTP)–Snurportin (PDB 3GJX), and a Saccharomyces cerevisiae 60S preribosomal particle (PDB 3JCT) are shown to scale with the human NPC symmetric core (41, 292–296). Abbreviations: NPC, nuclear pore complex; PDB, Protein Data Bank; POMs, pore membrane proteins.

Lin, D.H., Hoelz, A. (2019). Annu. Rev. Biochem. 88, 725-783
Figures from the paper:
Abstract:
The nuclear pore complex (NPC) serves as the sole bidirectional gateway of macromolecules in and out of the nucleus. Owing to its size and complexity (∼1,000 protein subunits, ∼110 MDa in humans), the NPC has remained one of the foremost challenges for structure determination. Structural studies have now provided atomic-resolution crystal structures of most nucleoporins. The acquisition of these structures, combined with biochemical reconstitution experiments, cross-linking mass spectrometry, and cryoelectron tomography, has facilitated the determination of the near-atomic overall architecture of the symmetric core of the human, fungal, and algal NPCs. Here, we discuss the insights gained from these new advances and outstanding issues regarding NPC structure and function. The powerful combination of bottom-up and top-down approaches toward determining the structure of the NPC offers a paradigm for uncovering the architectures of other complex biological machines to near-atomic resolution.
Structure of the Nuclear Pore Complex (An Update)

California Institute of Technology
Division of Chemistry & Chemical Engineering
1200 E. California Blvd.
Pasadena, CA 91125-7200
© Copyright Hoelz Laboratory

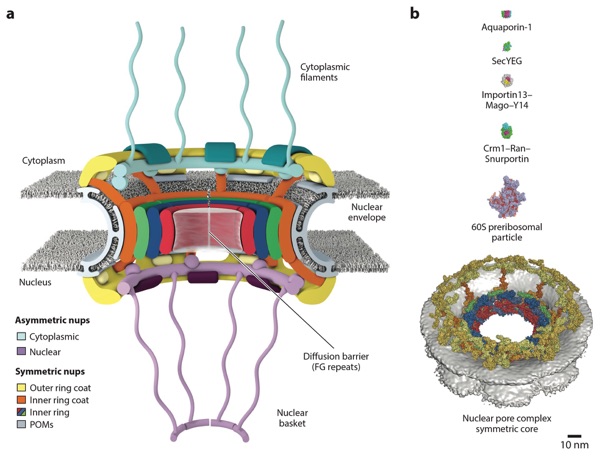
Figure 5. Structures of nucleoporins in the CNC. A schematic of the NPC is shown in the top right corner, with the region of the NPC in which these nucleoporins are found colored in yellow. Crystal structures of scSeh1–scNup85 (PDB 3F3F), scSec13–scNup145C–scNup84 (PDB 3IKO), Nup107–Nup133 (PDB 3I4R), spNup120–spNup37 (PDB 4FHN), Nup133 (PDB 1XKS), mmELYS (PDB 4I0O), and Nup43 (PDB 4I79) are shown in ribbon representation (64, 68, 70, 71, 73, 97). Abbreviations: ALPS, amphipathic lipid packing sensor; CNC, coat nucleoporin complex; DIM, domain invasion motif; NPC, nuclear pore complex; PDB, Protein Data Bank.
Figure 4. Domain architectures of human nucleoporins and summary of individual nucleoporins. Bars above domain architectures indicate the regions of the nucleoporins for which high-resolution structures are available, colored by the species from which the structures were determined. Abbreviations: CTD, C-terminal domain; NTD, N-terminal domain; nups, nucleoporin proteins; POMs, pore membrane proteins.
Figure 2. Progress in cryo-ET reconstructions of the NPC. Surface representations of cryo-ET reconstructions are shown to scale for Dictyostelium discoideum (EMD 1097), Homo sapiens (2012, EMD 5411; 2013, EMD 2444; 2015, EMD 3103), Chlamydomonas reinhardtii (EMD 4355), Xenopus laevis (EMDs 3005–3008), and Saccharomyces cerevisiae (EMD 7321) (31–33, 35, 36, 38–40). When possible, the nuclear envelope was colored yellow. For the D. discoideum reconstruction and the first H. sapiens reconstruction, the nuclear envelope is present but not colored. For the S. cerevisiae reconstruction, the NPCs were purified and the nuclear envelope is not present. Abbreviations: cryo-ET, cryo–electron tomography; EMD, Electron Microscopy Databank; NPC, nuclear pore complex.
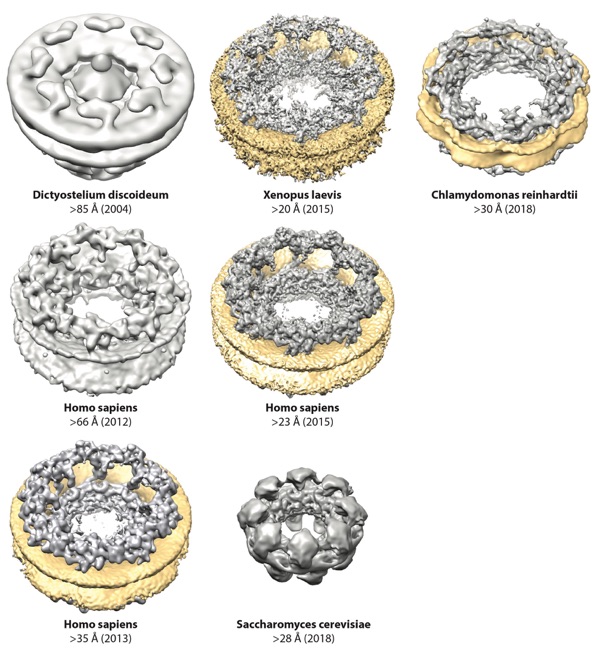
Figure 3. Nomenclature for nucleoporins in Saccharomyces cerevisiae, Chaetomium thermophilum, and Homo sapiens, grouped by subcomplex. Asterisks (∗) indicate that Nup98 and Nup62 and their homologs are also found in the cytoplasmic filaments or nuclear basket. The single dagger (†) indicates that ELYS is only a member of the nuclear coat nucleoporin complex in humans, rather than being symmetrically distributed. Its distribution in fungi remains uncertain. The double dagger (‡) indicates that Nup205/Nup188 molecules are also localized in the outer rings in humans. Stoichiometries of the nucleoporins in intact NPCs are indicated in parentheses next to the S. cerevisiae and H. sapiens nucleoporins (40, 50, 52). Abbreviations: NPC, nuclear pore complex; nups, nucleoporin proteins; POMs, pore membrane proteins.
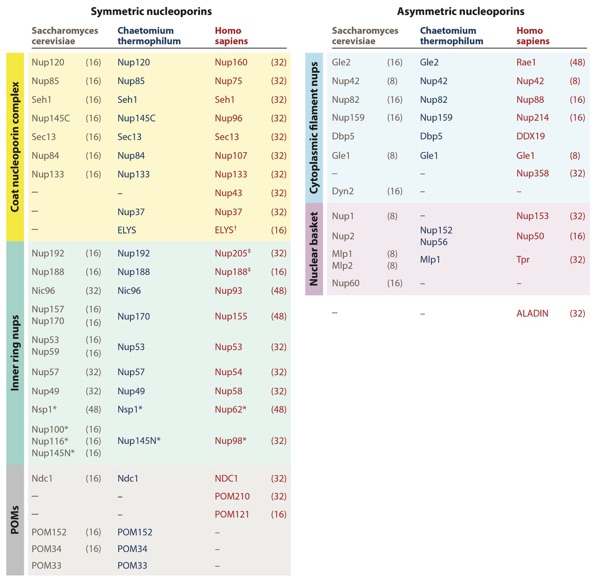

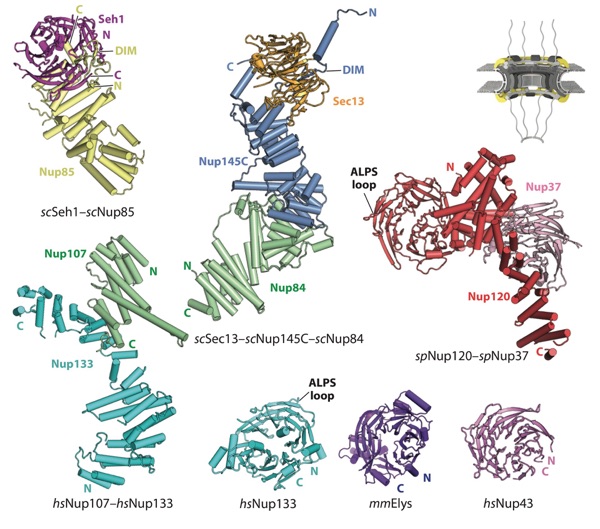
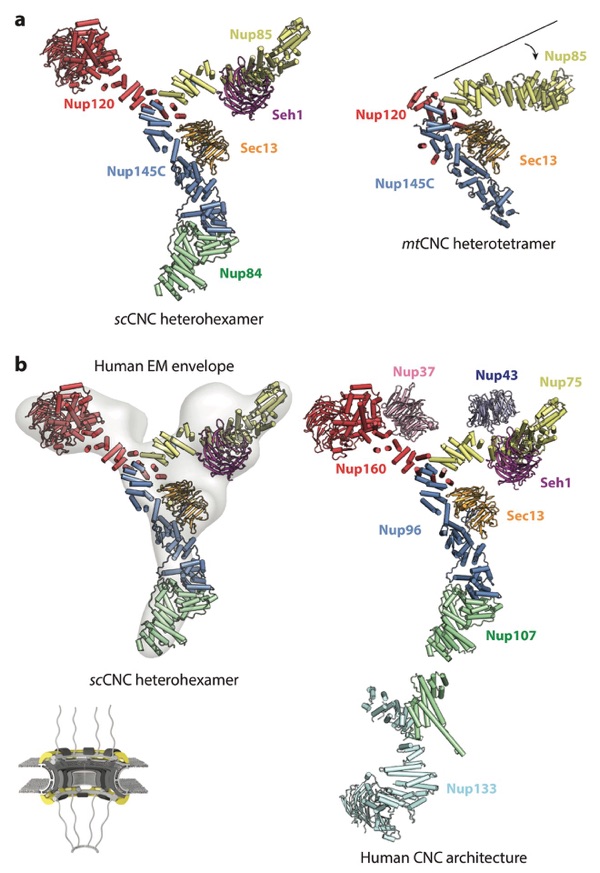
Figure 6. Architecture of the CNC. A schematic of the NPC is shown in the bottom left corner, with the region of the NPC in which these nucleoporins are found colored in yellow. Crystal structures of the (a) scSec13– scNup145C–scNup84–scSeh1–scNup85–scNup120 heterohexamer (PDB 4XMM) and mtSec13–mtNup145C– mtNup85–mtNup120 heterotetramer (PDB 4YCZ) are shown in the same orientation and coloring (75, 76). A crystallization chaperone antibody is omitted from the scCNC heterohexamer for clarity. Note the different angle that the Nup85 arm forms in the two structures. (b) A negative-stain electron tomography reconstruction of the human CNC (EMD 2443) is shown with the crystal structure of the scCNC heterohexamer fitted into it (38, 76). The arrangement of all subunits within the human CNC is shown in a composite structure obtained through fitting of individual CNC component structures into a cryo–electron tomographic reconstruction of the intact human NPC. Abbreviations: CNC, coat nucleoporin complex; EMD, Electron Microscopy Databank; NPC, nuclear pore complex; PDB, Protein Data Bank.
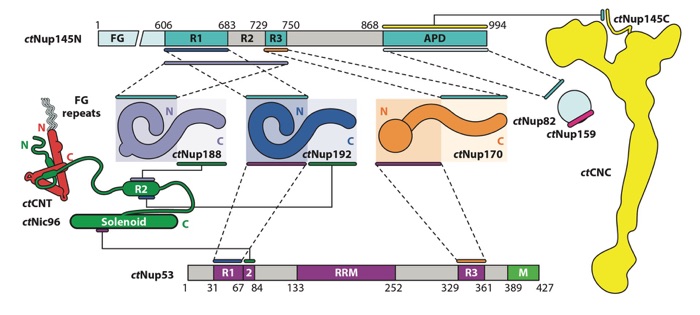
Figure 7. Biochemical interaction map for nucleoporins from the thermophilic fungus Chaetomium thermophilum. Lines or pairs of dashed lines connecting two nucleoporins indicate interactions mapped to the respective regions of those nucleoporins. The ctCNT and ctCNC, which are formed through large hydrophobic interfaces, are depicted as diagrams representing the experimentally observed shapes of the complexes rather than the individual nucleoporins. The ctNup82–ctNup159 and ctCNT–ctNic96 interactions are represented by diagrams of the interactions, rather than lines connecting the nucleoporins, as they have been characterized via X-ray crystallography. Figure adapted from Reference 41 with permission. Abbreviations: APD, autoproteolytic domain; CNC, coat nucleoporin complex; CNT, channel nucleoporin heterotrimer; RRM, RNA-recognition motif.
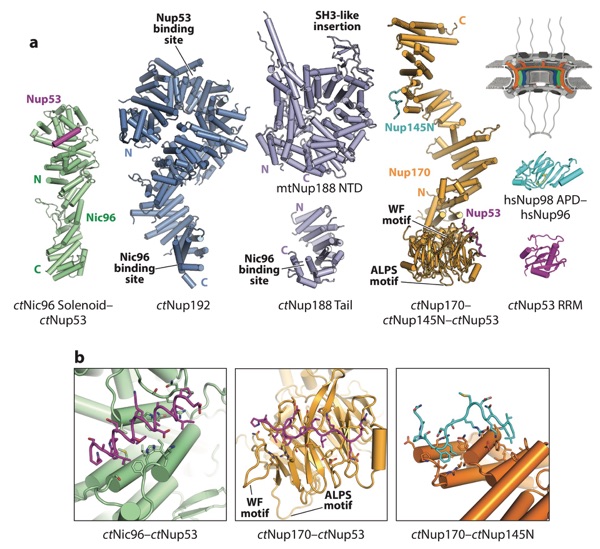
Figure 8. Structures of nucleoporins in the IRCs. A schematic of the NPC is shown in the top right corner, with the region of the NPC in which these nucleoporins are found colored in orange, green, and blue. (a) A crystal structure of the ctNic96–ctNup53 (PDB 5HB3), composite model of ctNup192 (derived from PDBs 4KNH, 5HB4, and 5CWV), model of ctNup188 (derived from PDB 4KF7 and 5CWU), composite model of ctNup170 (derived from PDBs 5HAX, 5HB0, and 5HB1), crystal structure of Nup98 bound to the N terminus of Nup96 (PDB 1KO6), and crystal structure of ctNup53 (PDB 5HB7) are shown in ribbon representation (41, 102). (b) Zoomed views of the ctNic96–ctNup53, ctNup170–ctNup53, and ctNup170–ctNup145N interactions. Figure adapted from Reference 41 with permission. Abbreviations: IRCs, inner ring complexes; NPC, nuclear pore complex; PDB, Protein Data Bank; RRM, RNA-recognition motif.
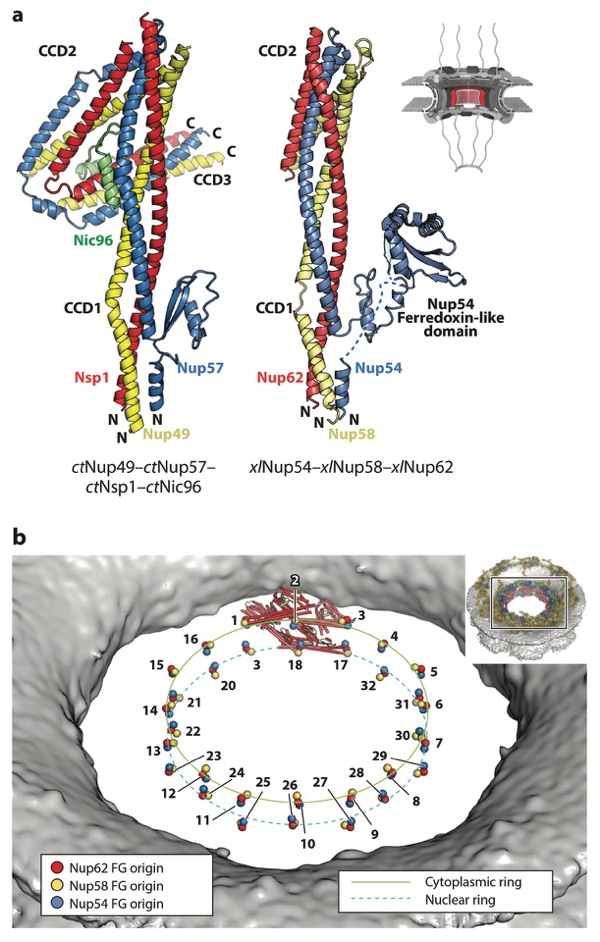
Figure 9. Structure of the CNT. A schematic of the NPC is shown in the top right corner, with the region of the NPC in which these nucleoporins are found colored in red. (a) Crystal structures of the ctNup49–ctNup57– ctNsp1–ctNic96 complex (PDB 5CWS) and a model of xlNup54–xlNup58–xlNup62 (combining PDBs 5C2U and 5C3L) are shown in ribbon representation in the same orientation and coloring (98, 113). Crystallization chaperone antibody (ctCNT–ctNic96) and nanobody (xlCNT) were omitted from both structures for clarity. In the xlNup54–xlNup58–xlNup62 complex, CCD3 was omitted for crystallization. The Nup54 ferredoxin-like domain is not present in fungi. (b) Positioning of CNTs in the human NPC maps the origins from which their N-terminal FG repeats project into the central transport channel to form the diffusion barrier (colored spheres). The CNTs from a single spoke are shown in ribbon representation. The FG origins form two stacked rings at the very center of the central transport channel. The inset shows the entire NPC, with a box indicating the orientation of the zoomed view. Abbreviations: CCD, coiled-coil domain; CNT, channel nucleoporin trimer; NPC, nuclear pore complex; PDB, Protein Data Bank.
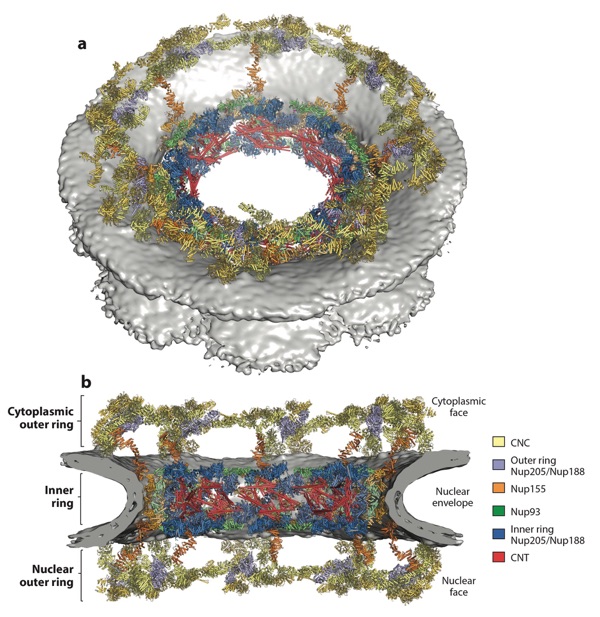
Figure 10. Composite structure of the human NPC symmetric core. (a) A view of the entire NPC from the cytoplasmic side with docked crystal structures shown in ribbon representation. The gray surface represents the nuclear envelope. (b) A cutaway of the NPC viewed from the plane of the nuclear envelope with docked crystal structures shown in ribbon representation. Because Nup188 and Nup205 possess very similar overall shapes and cannot be distinguished at current resolutions, the assignment of the six positions is tentative. Figure adapted from Reference 41 with permission. Abbreviations: CNC, coat nucleoporin complex; CNT, channel nucleoporin heterotrimer; NPC, nuclear pore complex.
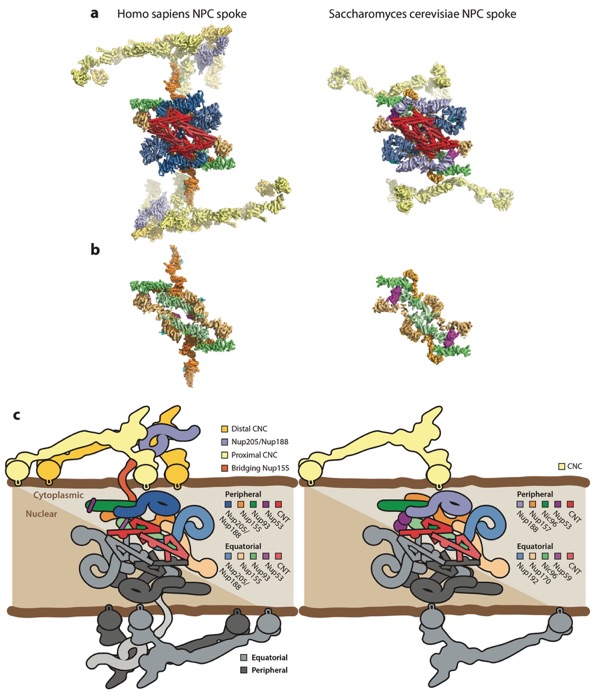
Figure 11. Arrangement of subunits within an NPC spoke. (a) The arrangement of subunits in one spoke from the human NPC symmetric core (left) and the arrangement of subunits from one spoke in the Saccharomyces cerevisiae NPC symmetric core (right) are shown from the same view in a surface view. (b) The same views as in panel a, but with the Nup205/Nup192, CNC, and CNT molecules removed to show the organization of molecules in the inner ring coat. (c) Schematics of the arrangement of subunits in the spokes. The precise orientation of the CNC in the S. cerevisiae NPC is ambiguous owing to the low resolution of the outer rings in the currently available reconstruction. Abbreviations: CNC, coat nucleoporin complex; CNT, channel nucleoporin heterotrimer; NPC, nuclear pore complex.
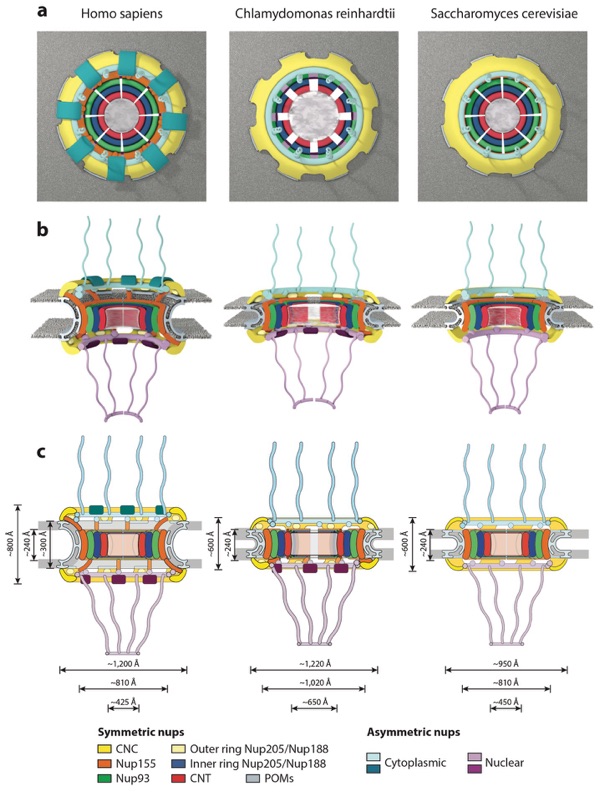
Figure 12. Conservation of NPC architecture. Schematics of the human, Chlamydomonas reinhardtii, and Saccharomyces cerevisiae NPC architectures are shown, depicting overall differences observed among the three species in the (a) top view, (b) side view, and (c) two-dimensional schematics. Notably, the S. cerevisiae NPC is smaller in height than the human NPC, is embedded in a thinner nuclear envelope, but has approximately the same size transport channel and inner ring complex. The C. reinhardtii NPC has a dilated inner ring and is embedded in a thinner nuclear envelope compared with the human NPC but has the same outer diameter. Abbreviations: CNC, coat nucleoporin complex; CNT, channel nucleoporin heterotrimer; NPC, nuclear pore complex; nups, nucleoporin proteins; POMs, pore membrane proteins.
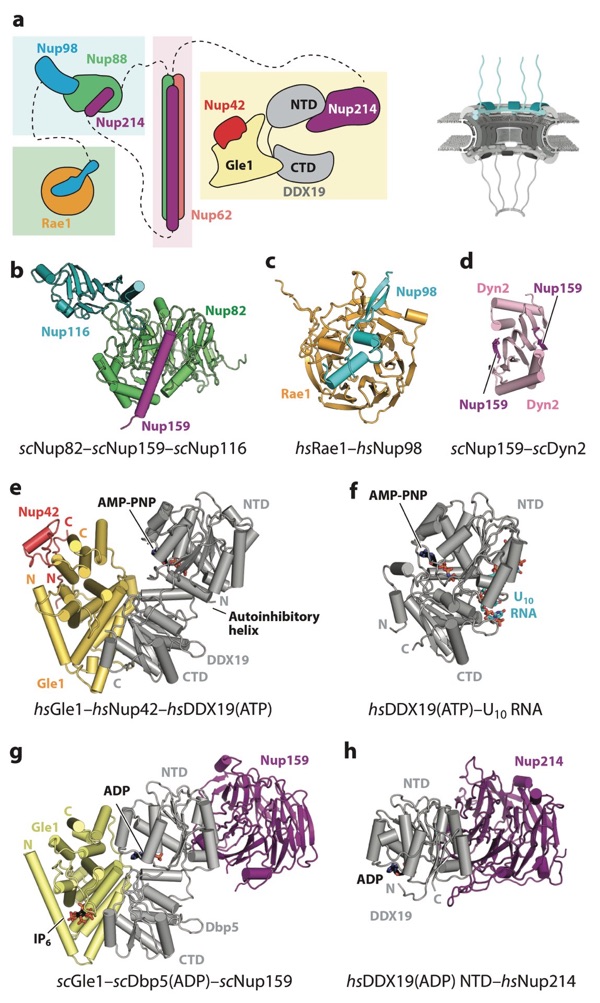
Figure 13. Structures of cytoplasmic filament nucleoporins. A schematic of the NPC is shown in the top right corner, with the region of the NPC in which these nucleoporins are found colored in cyan. (a) Schematic representation of the cytoplasmic filament interaction hubs. FG repeats and other regions are omitted for clarity. Dashed lines indicate nucleoporins that are connected to different interaction hubs via flexible linkers. (b) Crystal structure of the scNup82–scNup159–scNup116 complex (PDB 3PBP) shown in ribbon representation (126). (c) Crystal structure of the Rae1–Nup98 complex (PDB 3MMY) shown in ribbon re- presentation (171). (d) Crystal structure of the scNup159–scDyn2 complex (PDB 4DS1) shown in ribbon representation (175). (e) Crystal structure of the Gle1–Nup42–DDX19 complex (PDB 6B4J) shown in ribbon representation (177). Gle1 binds to DDX19 and Nup42 on opposite surfaces of the protein. (f) Crystal structure of the DDX19–U10 RNA complex (PDB 3G0H) shown in the same orientation as the Gle1–Nup42–DDX19 complex (205). DDX19 binds RNA and nucleotide on opposite surfaces of the protein. (g) Crystal structure of the scGle1–scDbp5–scNup159 complex (PDB 3RRM) shown in ribbon representation in the same orientation and coloring as the Gle1–Nup42–DDX19 complex (174). (h) Crystal structure of the DDX19–Nup214 complex (PDB 3FMP) shown in the same orientation as the scGle1–scDbp5–scNup159 complex (169). Abbreviations: AMP, adenylyl-imidodiphosphate; CTD, C-terminal domain; NPC, nuclear pore complex; NTD, N-terminal domain; PDB, Protein Data Bank.
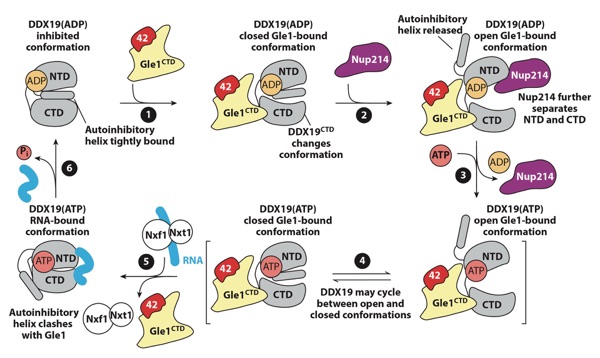
Figure 14. A schematic representation of the proposed regulatory cycle of the activation of DDX19 by the NPC. DDX19 resides in an autoinhibited conformation. (●1) Gle1 binding to the DDX19 CTD results in conformational rearrangements in the CTD that destabilize the autoinhibited conformation. (●2) Nup214 binding may promote a more open conformation of DDX19 and complete release from autoinhibition. (●3) Nucleotide exchange may be promoted while DDX19 is in a more open conformation. (●4) DDX19 is able to adopt the autoinhibited conformation in both ATP- and ADP-bound states and may cycle between the autoinhibited and closed conformations. (●5) Upon binding RNA, DDX19 adopts a closed, catalytically active state, and structural rearrangements are incompatible with Gle1 binding. (●6) ATP hydrolysis causes release of bound RNA and recycling of DDX19 to the beginning of the cycle. Figure adapted from Reference 177. Abbreviations: 42, Nup42; ADP, adenosine diphosphate; ATP, adenosine triphosphate; CTD, C-terminal domain; NPC, nuclear pore complex; NTD, N-terminal domain.
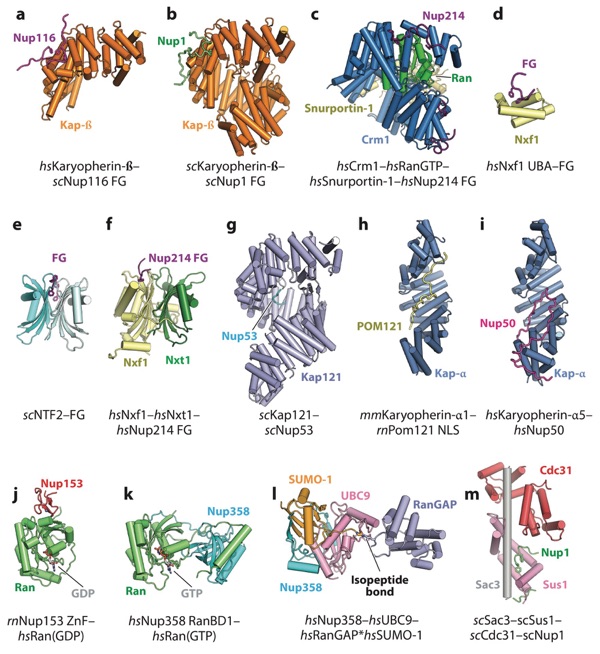
Figure 15. Structures of the interactions between the NPC and nucleocytoplasmic transport machinery. (a) Crystal structure of Karyopherin-β bound to an FG repeat from scNup116 (PDB 1O6P) (233). (b) Crystal structure of scKaryopherin-β bound to an FG repeat from scNup1 (PDB 5OWU) (240). (c) Crm1–Ran(GTP)–Snurportin-1 complex bound to several FG repeats from Nup214 (PDB 5DIS) (237). (d) Crystal structure of the Nxf1 UBA domain bound to the synthetic FG peptide (PDB 1OAI) (243). (e) Crystal structure of scNTF2 bound to a synthetic FG peptide (PDB 1GYB) (241). ( f ) Crystal structure of the Nxf1–Nxt1 complex bound to a FG repeat from Nup214 (PDB 1JN5) (244). (g) Crystal structure of scKap121 bound to an NLS-like peptide from scNup53 (PDB 3W3Y) (245). (h) Crystal structure of mmKaryopherin-α1 bound to the NLS from rnPom121 (PDB 4YI0) (246). (i) Crystal structure Karyopherin-α5 bound to the NLS-like sequence from Nup50 (PDB 3TJ3) (297). ( j) Crystal structure of the rnNup153 ZnF domain bound to Ran(GDP) (PDB 3GJ3) (224). (k) Crystal structure of the Nup358 RanBD1 bound to Ran(GTP) (PDB 1RRP) (214). (l) Crystal structure of the Nup358–UBC9–RanGAP∗SUMO-1 complex (PDB 1Z5S) (215). (m) Crystal structure of the scSac3–scSus1–scCdc31 TREX-2 complex bound to an FG-like sequence from scNup1 (PDB 4MBE) (258). Abbreviations: NLS, nuclear localization signal; NPC, nuclear pore complex; PDB, Protein Data Bank; RanBD, Ran-binding domain; UBA, ubiquitin-associated; ZnF, zinc finger.
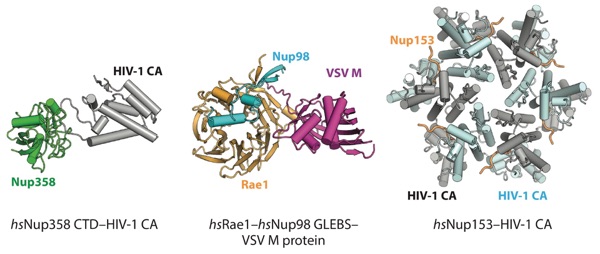
Figure 15. Structures of interactions between the NPC and viral factors. Crystal structures of the Nup358–HIV-1 CA protein complex (PDB 4LQW), Rae1–Nup98 GLEBS–VSV M protein complex (PDB 4OWR), and Nup153–HIV-1 CA protein complex (PDB 4U0C) are shown in ribbon representation (176, 288, 290). Abbreviations: CA, capsid protein; CTD, C-terminal domain; GLEBS, Gle2-binding sequence; NPC, nuclear pore complex; PDB, Protein Data Bank; VSV, vesicular stomatitis virus.
