Hoelz Lab: Publications
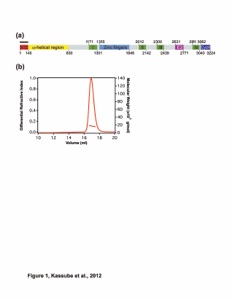
Figure 1. (a) Spatial organization of nucleoporin classes, as listed in Table 1. (b) Crystal structure of the fully assembled S. cerevisiae CNC4, shown in cartoon representation (PDB 4XMM; lacking Nup133). The CNC crystal structure reveals the overall conformation of the CNC and the molecular details of how the three different arms of the Y-shaped CNC form the central triskelion. The conformation-specific high-affinity synthetic antibody sAB-57 (gray) was used as a crystallization chaperone and yielded essential crystal-packing interactions. NTD, N-terminal domain. (c,d) The S. cerevisiae CNC crystal structure fitted into the negative-stain EM reconstructions of the S. cerevisiae (c) and human (h) CNC (d). The CNC crystal structure and EM envelopes are illustrated in cartoon representation and gray transparent surfaces, respectively. Notably, additional density for the two human coat nucleoporins Nup37 and Nup43 is observed in the EM reconstruction of the human CNC. Images in a–d are adapted with permission from ref. 4, AAAS. (e) The negative-stain EM structure of the isolated human CNC (also shown in d) and the tomographic map of the entire NPC, resolved to ~32 Å (EMD-2444 and EMD-2443)3. A systematic fitting analysis revealed that 32 copies of the CNC fit into 16-membered nucleoplasmic and cytoplasmic CNC rings (only one is shown fully) of the tomographic map (transparent gray). The fitting suggests the formation of two concentric, reticulated eight-membered rings on both the nucleoplasmic and cytoplasmic sides of the NPC, in which the CNCs are arranged in a head-to-tail fashion. The outer (blue) and inner (green) CNC rings are rotated slightly with respect to each other. The nuclear envelope is illustrated in yellow. Adapted with permission from ref. 3, Elsevier.

Figures from the paper:
Abstract:
Elucidating the structure of the nuclear pore complex (NPC) is a prerequisite for understanding the molecular mechanism of nucleocytoplasmic transport. However, owing to its sheer size and flexibility, the NPC is unapproachable by classical structure determination techniques and requires a joint effort of complementary methods. Whereas bottom-up approaches rely on biochemical interaction studies and crystal-structure determination of NPC components, top-down approaches attempt to determine the structure of the intact NPC in situ. Recently, both approaches have converged, thereby bridging the resolution gap from the higher-order scaffold structure to near-atomic resolution and opening the door for structure-guided experimental interrogations of NPC function.
California Institute of Technology
Division of Chemistry & Chemical Engineering
1200 E. California Blvd.
Pasadena, CA 91125-7200
© Copyright Hoelz Laboratory

Toward the atomic structure of the nuclear pore complex: when top down meets bottom up
Hoelz, A.* Glavy, J.* Beck, M.*
(2016). Nature Structural & Molecular Biology. 23, 624-630
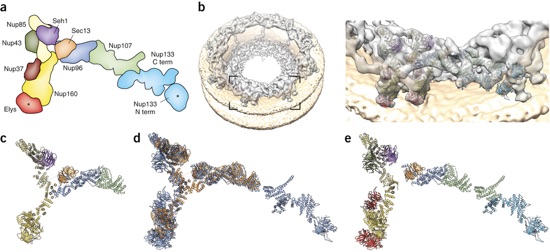
Figure 2. (a) Schematic representation of the vertebrate CNC, with the positions of all components indicated. The human coat nucleoporins Nup160, Nup96 and Nup107 correspond to yeast coat nucleoporins Nup120, Nup145C and Nup84 (Fig. 1b), respectively. The human coat nucleoporins Nup43, Nup37 and Elys are absent in the S. cerevisiae CNC. Asterisks mark domains that can be confidently positioned on the basis of available data but not accurately oriented, owing to a lack of structural data. Term, terminus. Adapted from ref. 34, Nature Publishing Group. (b) Tomographic map of the human NPC at a resolution of up to ~23 Å. The inset indicates a rotational segment of the nuclear ring that is magnified on the right with the fitted coat nucleoporin crystal structures shown in cartoon representation, colored according to a. (c) Crystal structure of the S. cerevisiae CNC (lacking Nup133). (d) Superposition of the structures in c (displayed in orange) and e (displayed in blue). Both structures were not fitted to each other but are shown superimposed on the basis of independent fitting into the tomographic map. The superposition therefore effectively illustrates the accuracy of the present structural assignments. (e) Hybrid structure obtained independently by fitting the individual X-ray structures into the tomographic map.
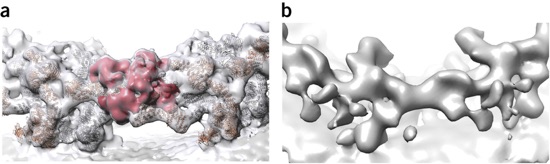
Figure 3. (a,b) Rotational segment (asymmetric unit) of the cytoplasmic ring of the human NPC obtained from Nup358 gene-silenced cells (b) or untreated cells (a, putative Nup358 density segmented in red). The electron optical density of the outer CNC is absent after gene silencing of Nup358 in the case of the cytoplasmic ring but not the nuclear ring (not shown). Adapted from ref. 34, Nature Publishing Group.
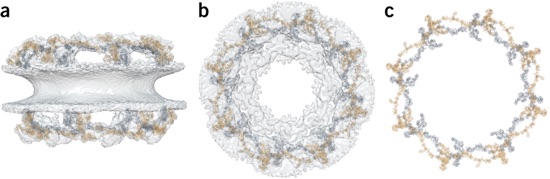
Figure 4. Details are discussed in the text. (a) Side view of the nuclear pore, as observed by cryo-EM, superposed with the inner (gray) and outer (orange) Y complexes. (b) As in a, but in top view. (c) As in b, but without any EM density.
