Hoelz Lab: Publications
Figure 1. The 6D7A loop of the Nup214 NTD is essential for Ddx19 binding. (A) Domain organization of Nup214 and Ddx19. For Ddx19, the N-terminal extension (NTE, orange) and the 2 RecA-like domains (domain 1 and 2, green and light orange) are indicated. For Nup214, the β-propeller domain (light blue) and its C-terminal extension (CTE, yellow), the coiled-coil domain (gray), and the C-terminal unstructured region containing numerous FG-repeats (white) are indicated. The Nup214 NTD is composed of the β-propeller domain followed by the CTE. The bars above the domain structures mark the fragments of the orthorhombic crystal form. (B) Gel filtration profiles of full-length wild-type Ddx19 incubated with the Nup214 NTD, Nup214 NTD 1–405, or Nup214 NTD Δ6D7A before injection.

Napetschnig, J., Kassube, S.A., Debler, E.W., Wong, R.W., Blobel, G. and Hoelz, A.
(2009). Proc. Natl. Acad. Sci. USA 106, 3089-3094.
Figure 2. Overview of the Nup214 NTD·Ddx19 NTD structure. Ribbon representation of the Nup214 NTD·Ddx19 NTD complex (Upper). A 90° rotated view is shown in Lower. For the Nup214 NTD, the β-propeller domain (blue), the 6D7A loop (magenta), the C-terminal extension (CTE; yellow), and the blade numbers are indicated. For the Ddx19 NTD, the N-terminal RecA-like domain (green) and the unique N-terminal extension (NTE; orange) is indicated. The ADP molecule bound to the Ddx19 NTD is shown in ball-and-stick representation.
Figure 3. Surface properties of the Nup214 NTD-Ddx19 NTD interaction. (A) Surface renditions of the NTDs of Nup214 and Ddx19 in an open book representation colored according to the participation of the various domains as in Fig. 2. Surfaces that mediate the association between the 2 proteins are indicated in green (Ddx19) and blue (Nup214). As a reference, a ribbon representation of the complex is shown in its original orientation. (B) Surface representation colored according to a multispecies sequence alignment (Fig. S1). The conservation at each position is mapped onto the surface and is shaded in a color gradient from light yellow (60% similarity) to dark red (100% identity). (C) Surface representation colored according to the electrostatic potential. The electrostatic potential is plotted onto the surface and colored in a gradient from red (−10 kBT/e) to blue (+10 kBT/e). The orientation of all surface representations is identical. Black lines indicate the interface borders.
Figure 4.The conserved arginine 259 of Ddx19 is a key residue for complex formation. (A) Details of the interaction between the NTDs of Nup214 and Ddx19. The ribbon representation is colored according to Fig. 2. The Inset illustrates the position of R259 and its interacting residues and is expanded on the right. (B) Multispecies sequence alignment of Ddx19 homologs. The red asterisk indicates the location of the invariant R259. The conserved sequence motifs II and III are highlighted in gray boxes. Invariant kBT residues outside of the conserved sequence motifs and R259 are illustrated in red. The residue numbering is relative to human Ddx19 and the secondary structure of Ddx19 is shown above the sequence alignment.
Figure 5. A 9-residue region in the 6D7A loop of the Nup214 NTD is essential for Ddx19 binding. Gel filtration profiles of full-length wild-type Ddx19 incubated with the deletion mutants Nup214 NTDΔ1, Δ2, or Δ3 before injection. Gel filtration profiles of Ddx19 are colored in gray, the Nup214 NTD variants in blue, and the elution profile resulting from incubating Ddx19 with Nup214 proteins in red. The red arrow indicates the expected elution volume of the Nup214·Ddx19 complex (see Fig. 1B Top).
PDB coordinates (link to PDB site) - Space Groups P21, P212121
PDB coordinates - Space Groups P21 (.pdb), P212121 (.pdb)
Structure factors - Space Groups P21 (.txt), P212121 (.txt)
Figures from the paper:
Coordinates:
Abstract:
Key steps in the export of mRNA from the nucleus to the cytoplasm are the transport through the nuclear pore complex (NPC) and the subsequent remodeling of messenger RNA-protein (mRNP) complexes that occurs at the cytoplasmic side of the NPC. Crucial for these events is the recruitment of the DEAD-box helicase Ddx19 to the cytoplasmic filaments of the NPC that is mediated by the nucleoporin Nup214. Here, we present the crystal structure of the Nup214 N-terminal domain in complex with Ddx19 in its ADP-bound state at 2.5 A resolution. Strikingly, the interaction surfaces are not only evolutionarily conserved but also exhibit strongly opposing surface potentials, with the helicase surface being positively and the Nup214 surface being negatively charged. We speculate that the positively charged surface of the interacting ADP-helicase binds competitively to a segment of mRNA of a linearized mRNP, passing through the NPC on its way to the cytoplasm. As a result, the ADP-helicase would dissociate from Nup214 and replace a single bound protein from the mRNA. One cycle of protein replacement would be accompanied, cooperatively, by nucleotide exchange, ATP hydrolysis, release of the ADP-helicase from mRNA and its rebinding to Nup214. Repeat of these cycles would remove proteins from a mRNP, one at a time, akin to a ratchet mechanism for mRNA export.
Figure 6. Arginine 259 of Ddx19 is crucial for binding to Nup214 NTD. Gel filtration profiles of the Nup214 NTD (blue), the Ddx19 mutants R259A, R259Q, R259K and R259E (gray), and the elution profile resulting from incubation of Nup214 NTD with Ddx19 (red) before injection are indicated. The red arrow indicates the expected elution volume of the Nup214·Ddx19 complex (see Fig. 1B Top).
Figure 7. In vivo localization of Ddx19 and Ddx19 mutants. HeLa cells were transfected with Ddx19 and Ddx19 mutants containing a C-terminal HA-tag and analyzed with confocal microscopy. The monoclonal antibody mAb414 was used as a reference for nuclear envelope staining (red). The cellular localization of HA-tagged proteins was detected with an anti-HA antibody (green). The merged image reveals the colocalization of wild type Ddx19 and Ddx19 R259K with the nuclear envelope, while Ddx19 R259A displays no detectable nuclear envelope staining.
Figure S1. Multispecies sequence alignment of Ddx19 homologs. The overall sequence conservation at each position is shaded in a color gradient from yellow (60% similarity) to dark red (100% identity), using the Blosum62 weighting algorithm. The secondary structure of Ddx19 as observed in the Nup214 NTD•Ddx19 structure is shown above the alignment. The residue numbering is relative to H. sapiens Ddx19 and gray boxes below the alignment indicate the position of the 9 conserved DEAD-box helicase motifs. As a reference, the primary sequence of the M. janaschii DEAD-box protein MjDEAD is aligned with the Ddx19 homologs.
Figure S2. Salt dependence of the Nup214 –Ddx19 interaction. Gel filtration profiles of the Nup214•Ddx19 complex at identical protein complex concentrations of 50 uM and the indicated salt concentrations. The elution positions for molecular weight standards are indicated.
Figure S3. Comparison of the 6D7A loops of the human Nup214 and the yeast Nup159 β-propeller domains. (A) Structural alignment of the 6D7A loops of the homologous human and yeast β-propellers. The surface of the Nup214 β-propeller domain omitting the 6D7A loop is shown in a gray surface rendition. The Calpha-traces of the 6D7A loop of Nup214 (red) and Nup159 (blue) are shown in coil representation and side chains in ball-and-stick representation. For clarity, only the Nup214 residues are numbered. The localizations of the Nup159 mutants V323E and I326E that abolish complex formation with the yeast Dbp5 helicase are indicated by green dots. (B) Structure-based sequence alignment of the Nup214 and Nup159 6D7A loops. The secondary structure and residue numbering are indicated above and below the sequence for Nup214 and Nup159, respectively. The localization of the Nup159 double mutant V323E/I326E is labeled by green dots, and conserved residues are highlighted. The residues of the 6D7A loop that are deleted in the Nup214 NTD Δ1, Δ2, and Δ3 variants are indicated.


Figure S4. Analysis of the Nup214•Ddx19 interaction by peptide array. (A) Ddx19 peptide array probed with the radioactively labeled Nup214 NTD. The 5 identified regions containing at least 4 consecutive peptide spots are numbered I to V and are indicated by a red box. (B) Domain structures of Ddx19 and the M. jannaschii DEAD-box protein MjDEAD, displaying the 9 conserved sequence motifs as indicated in Fig. S1. The 5 identified regions and their corresponding residues are indicated. (C) Homology model of Ddx19 in surface representation based on MjDEAD (PDB code 1HV8) (3). Identified peptides were mapped onto the surface and colored in red. The residue numbering is relative to human Ddx19.
Figure S5. Alanine-scanning mutagenesis of Ddx19. Homology model of Ddx19 illustrating the approximate location of mutated Ddx19 surface residues. Alanine mutants that were capable of interacting with Nup214 are shown in yellow, and mutant R259A, which is incapable of interacting with Nup214, is shown in red. The residue numbering is relative to human Ddx19.

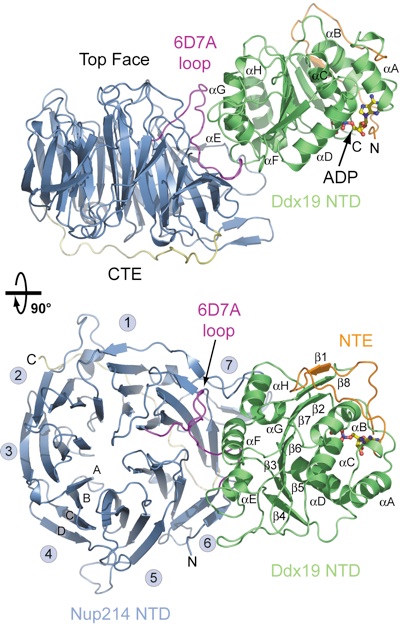
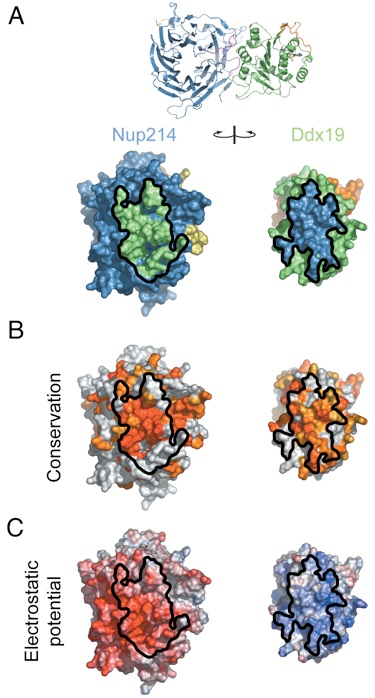

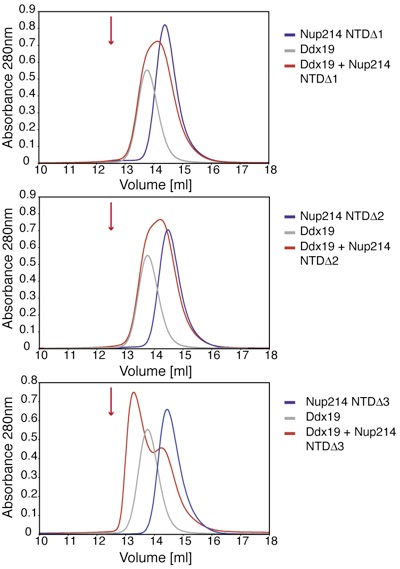

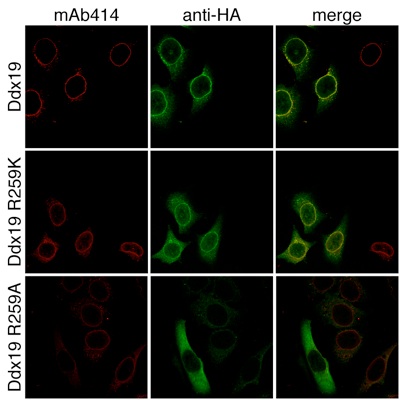
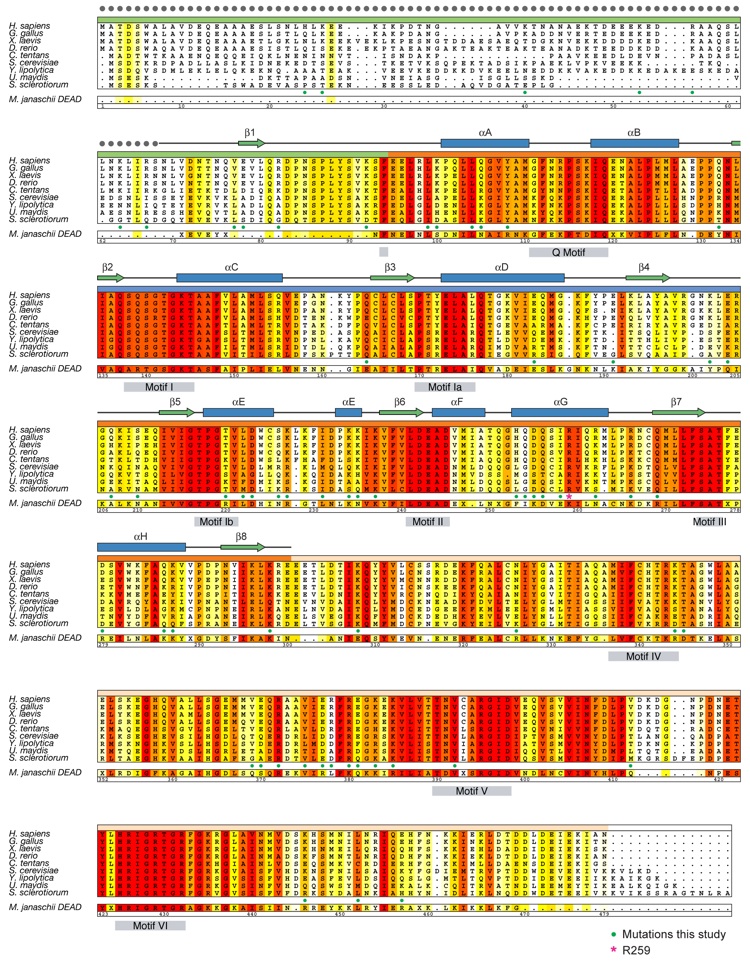
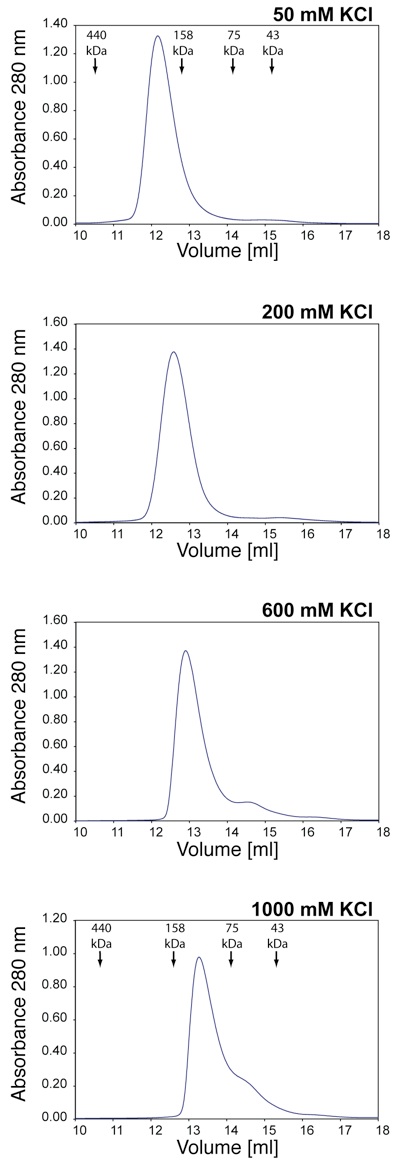
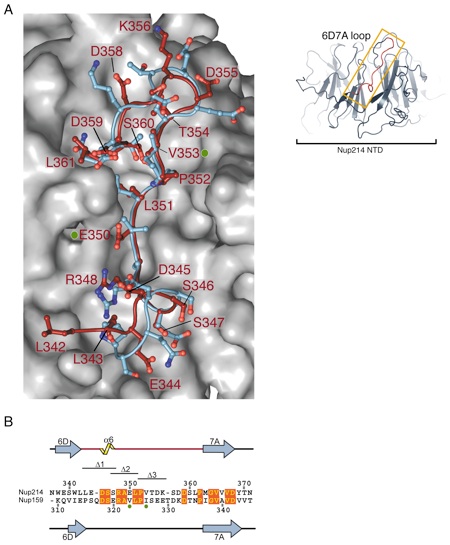
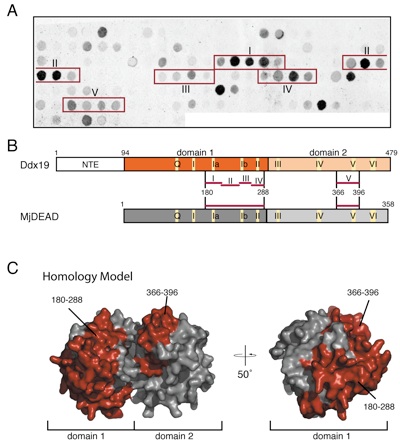

Figure S6. In vivo localization of Nup214 and Nup214 fragments. (A) The domain organization of Nup214 and the various Nup214 fragments are indicated. (B) HeLa cells were transfected with Nup214 and Nup214 fragments containing a C-terminal HA-tag and analyzed with confocal microscopy. The cellular localization of the HA-tagged Nup214 proteins was detected with an anti-HA antibody (green). The polyclonal anti-Nup358 antibody was used as a reference for nuclear envelope staining (red). The merged image is shown on the right.

Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19.
California Institute of Technology
Division of Chemistry & Chemical Engineering
1200 E. California Blvd.
Pasadena, CA 91125-7200
© Copyright Hoelz Laboratory
