Hoelz Lab: Publications
Figure 1. Domain organization and structure of Nup358CTD. (a) Domain organization of human Nup358. Domain boundaries are indicated by residue numbers. The bar above the domain structure denotes the crystallized fragment. I, II, III, and IV, Ran binding domains; NTD, N-terminal domain; CTD, C-terminal domain; E3, E3 ligase domain. (b) Cartoon representation of Nup358CTD with a view rotated 180° along the vertical axis shown on the right. (c) Cartoon representation of Nup358CTD rotated 90° along the horizontal axis from above. (d) Representative 2|Fo|-|Fc| electron density map contoured at 1.5 σ.

Figure 2. Surface properties of Nup358CTD. (a) Surface representation of Nup358CTD, with the active site colored in red. (b) Surface representation colored according to sequence identity based on a multi-species sequence alignment (Fig. S2). The identity at each position is mapped onto the surface and is shaded in a color gradient from white (60 % less than 60 % identity) to red (100 % identity). (c) Surface representation colored according to electrostatic potential from -10 kBT/e (red) to +10 kBT/e (blue).
Figure 3. Comparison of Nup358CTD and Cyclophilin A active sites. (a) Detailed view of the Nup358CTD active site. (b) Detailed view of the Cyclophilin A active site (PDB Code 1M9C). (c) Overlay of the active sites from Nup358CTD and Cyclophilin A. Critical active site residues are shown in stick representation, and the Cα-traces are shown in coil representation, according to the coloring scheme in A. The orientation of all active sites is identical.
Figure 4. Nup358CTD possesses peptidyl-prolyl isomerase activity. (a) Michaelis-Menten plot of the peptidyl-prolyl isomerization of Suc-Ala-Ala-Pro-Phe-2,4-difluoroanilide by Cyclophilin A, Nup358CTD, and Nup358CTD V3173W. (b) Michaelis-Menten plot of the peptidyl-prolyl isomerization of Suc-Ala-Ala-Pro-Phe-2,4-difluoroanilide by Nup358CTD and Nup358CTD V3173W. Note the different scale of the y-axis from panel (a). (c) Representative time-course traces of the peptidyl-prolyl isomerization of Suc-Ala-Ala-Pro-Phe-2,4-difluoroanilide by Cyclophilin A, Nup358CTD, Nup358CTD V3173W, and in the absence of an enzyme.
Figure 5. Structural comparison of Nup358CTD to the Cyclophilin A●HIV-1CA complex. The structure of Nup358CTD overlaid on the structure of the Cyclophilin A●HIV-1CA complex (PDB code 1M9C). The right panel is a close-up view of the interaction with the HIV-1CA loop rotated 90° along the vertical axis from the left panel. Note the clash between the Nup358CTD Q3163 and the HIV-1CA proline-rich loop.
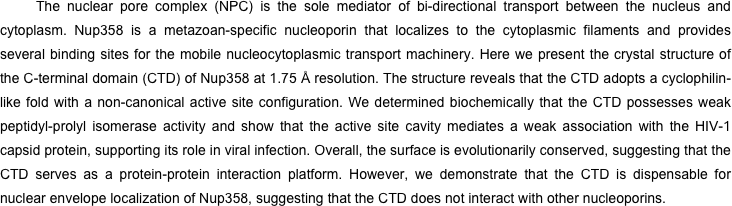
Figure S1. Multi-angle light scattering analysis of Nup358CTD. The differential refractive index and determined molecular weight are plotted against elution volume off of a Superdex 75 10/300 GL gel-filtration column.
Figures from the paper:
Coordinates:
Abstract:
California Institute of Technology
Division of Chemistry & Chemical Engineering
1200 E. California Blvd.
Pasadena, CA 91125-7200
© Copyright Hoelz Laboratory

Figure 6. Nup358CTD binds weakly to the HIV-1 capsid protein. (a-c) Size exclusion chromatography interaction analysis of HIV-1CA with (a) Cyclophilin A, (b) Nup358CTD, and (c) Nup358CTD V3173W. The analyzed fractions are indicated in gel-filtration profile by a grey bar. Notably, experiments were carried out with identical protein concentrations, the different peak height of the wild-type Nup358CTD is a result of a lower absorbance coefficient.
Figure 7. Nup358CTD is dispensable for nuclear envelope localization. Nup358CTD and Nup358 fragments carrying a N-terminal HA-tag were transiently transfected into HEK293T cells and analyzed by fluorescence microscopy. HA-tagged Nup358 protein localization was detected with an α-HA antibody (green). The monoclonal α-mAb414 antibody (red) and DAPI (blue) were used as a reference for nuclear envelope and nucleus staining, respectively. The right panel shows the merged images.
Structural and Functional Analysis of the C-terminal Domain of Nup358/RanBP2
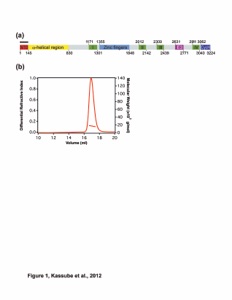

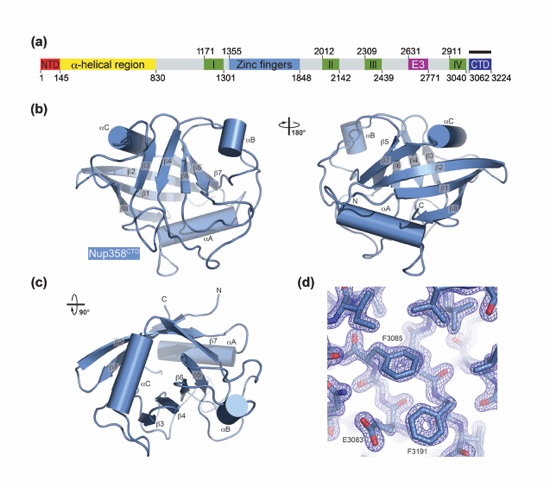




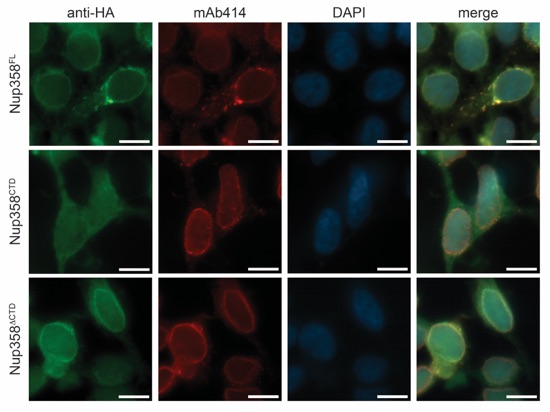

Figure S2. Multi-species sequence alignment of Nup358CTD. The overall sequence identity is shaded from white (less than 60 % identity) to red (100 % identity). The secondary structure as observed in the Nup358CTD structure is shown above the alignment, with blue bars representing α helices and green arrows representing β strands. The sequence of human cyclophilin A is displayed at the bottom of the alignment with residues identical to human Nup358CTD shaded in grey. Gray and magenta dots indicate identical and different active site residues between human Nup358CTD and human cyclophilin A, respectively. The numbering below the sequence is relative to human Nup358.
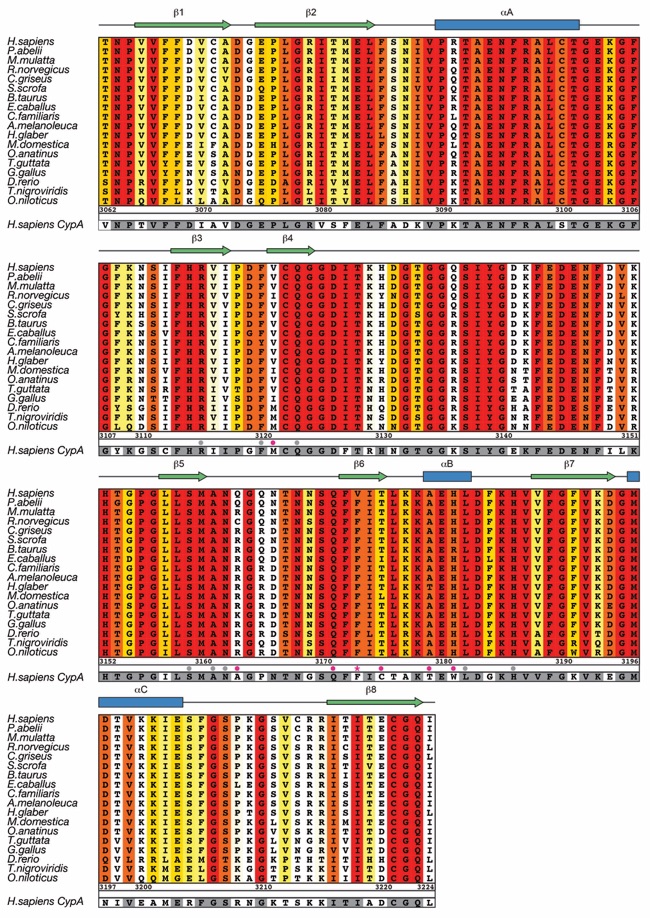
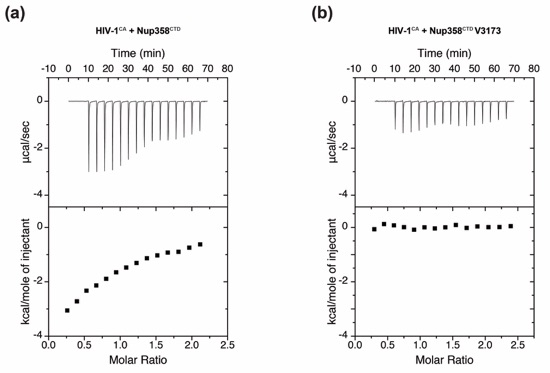
Figure S3. Isothermal titration calorimetry (ITC) analysis. Upper parts of each box show raw data and lower parts show integrated heat changes corrected for heat from dilution for interactions between (a) Nup358CTD and HIV-1CA and (b) Nup358CTD V3173W and HIV-1CA.

Lin, D.H., Zimmermann, S., Stuwe, T.S., Stuwe, E., Hoelz, A.*
(2013). J. Mol. Biol. 425, 1318-1329