Hoelz Lab: Publications
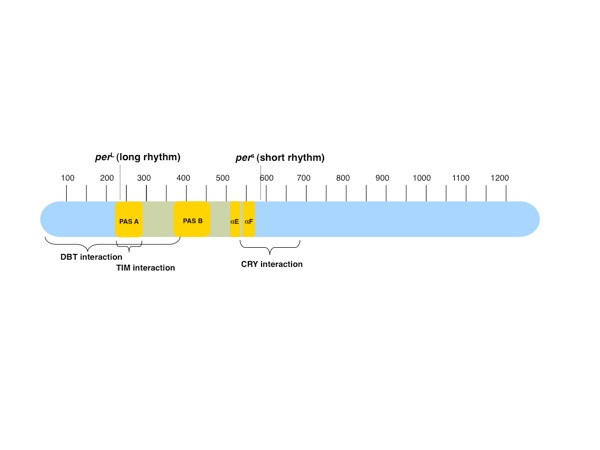
Figure 1. The PAS domain region of PER. The 1224-residue PER protein contains two PAS domains (PAS-AB) adjacent to one another in the N-terminal half of the protein. A classic PER allele, PER long (perL), is caused by the point mutation V243D in PAS-A. Following the PER PAS domain region is another classic allele, PER short (pers), caused by the mutation S589N in the short mutable region (residues 585–600) where phosphorylation events lead to degradation. Interactions between PER and the clock proteins DBT, TIM, and cryptochrome (CRY) have been mapped to regions surrounding and overlapping the PER PAS domain region.

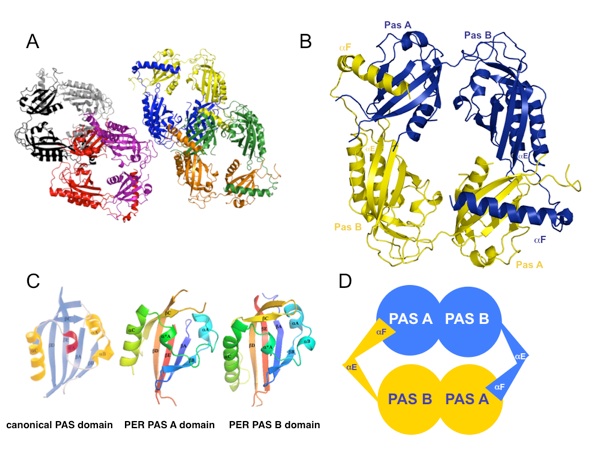
Figure 2. The PER PAS domain structure. (a) Four PER PAS-AB dimers are found in the asymmetric unit (black/gray, red/magenta, blue/yellow, and orange/green). (b) Each molecule includes two PAS domain regions (PAS-A and PAS-B) followed by two additional helices. The final helix (αF) of one molecule wraps around the dimer to pack against the PAS-A β-sheet of the opposing subunit. (c) The PAS domains of PER assume a canonical PAS fold with secondary structure labeled accordingly (βA, βB, αA, αA⁎, αB, αC, βB, βC, and βD). (d) A schematic representation of the PAS-AB symmetric dimer.
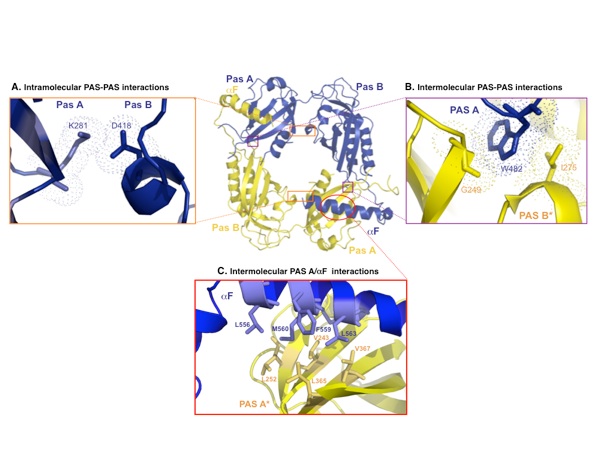
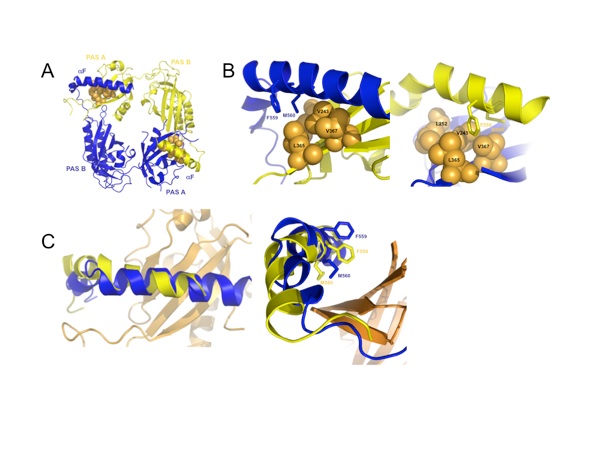
Figure 3. Interactions between the PAS subunits. (a) Intramolecular interactions between the PAS-A and the PAS-B domains. Lys281 of PAS-A forms a salt bridge with Asp418 at the interface between the PAS-A and the PAS-B domains of each molecule. Dots represent van der Waals surfaces. (b) Intermolecular interactions occur between Trp482, which protrudes from the PAS-B domain of the first molecule, and Gly248 and Ile275 on the PAS-A domain of the second molecule. (c) Intermolecular interactions at the PAS-A–αF interfaces. Each αF that reaches out forms a hydrophobic interface with the PAS-A β-sheet of the partner molecule.
Figure 4. Each of four crystallographic homodimers of the asymmetric unit exhibits two distinct positions for αF in the PAS-A–αF interaction between opposing subunits. (a) The position of αF relative to the hydrophobic PAS-A β-sheet differs in the two subunits (key interface residues are Leu252, Val243, Leu365, and Val367, shown in orange). (b) A twist of αF between the two helical positions directs either Met560 (blue, left) or Phe559 (yellow, right) into the center of the hydrophobic patch. (c) An overlay of the two F helices demonstrates their different positions relative to PAS-A β-sheet (orange).
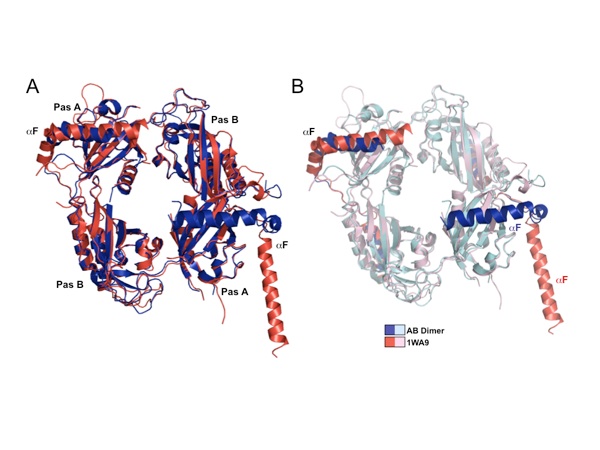
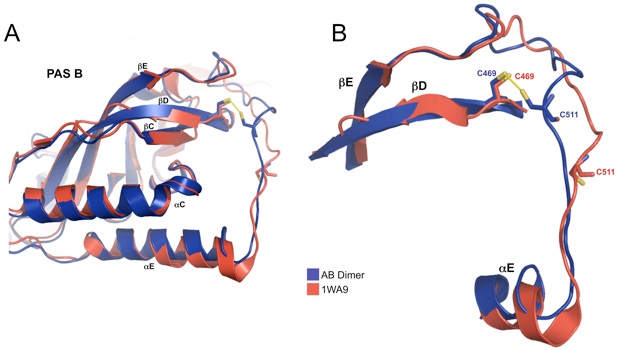
Figure 5. Release of αF from the PAS domain core (1WA9). (a) Compared to a previously determined PER structure (1WA9), the new structure (blue) shows an alternative conformation of αF, which assumes a closed position in both molecules rather than one closed helix and one extended helix as in 1WAP (red) (b) The major differences between the two structures involve the αF positions.
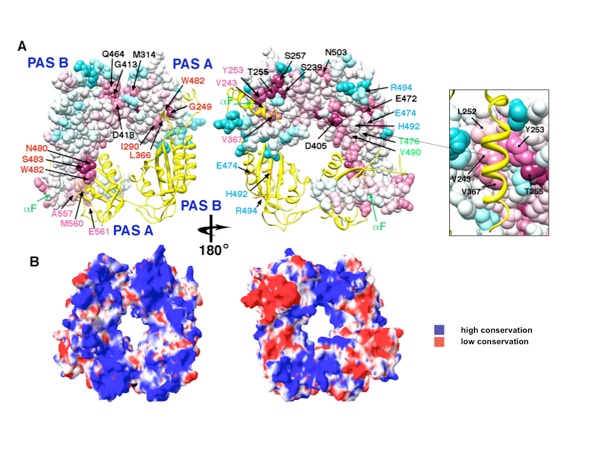
Figure 6. Conservation of surface residues and electrostatic properties of the PER dimer. (a) Conservation of residue type displayed on the PER dimer (one subunit shown as a ribbon, the other, as a van der Waals surface). Residues are colored based on degree of conservation from an alignment of 28 related PAS-AB proteins (e.g., PER, mPER, CLK, ARNT, HiF-1α, BMAL, and NPAS2) with an overall sequence identity greater than 22% [scale ranges from invariant positions (dark purple) to variable (blue)]. For sequence alignment, see Supplemental Data. Residues at the PAS-AB contact are highly conserved, as those that mediate the αF and PAS-A⁎ interaction (pink labels). Another pocket of conservation includes residues on PAS-A that accept W482 from PAS-B (red labels). Other conserved patches (black labels) have no currently known function. Residues implicated in TIM binding on PAS-B (blue labels) are not conserved among PAS-AB family members and neither are those that mediate the PAS-B–PAS-B contact in the dimer of mPER (blue and green labels). Conservation mapping performed with ConSurf and rendered in Chimera. (b) The PER dimer has an asymmetric distribution of electrostatic potential, with one face more positive than the other. Electrostatic potential calculated with the Poisson–Boltzmann equation and mapped on to the solvent-accessible surface of PER (blue = 2 kT/q, red = − 2kT/q; internal protein dielectric = 4.0; partial charges assigned to protein atoms).
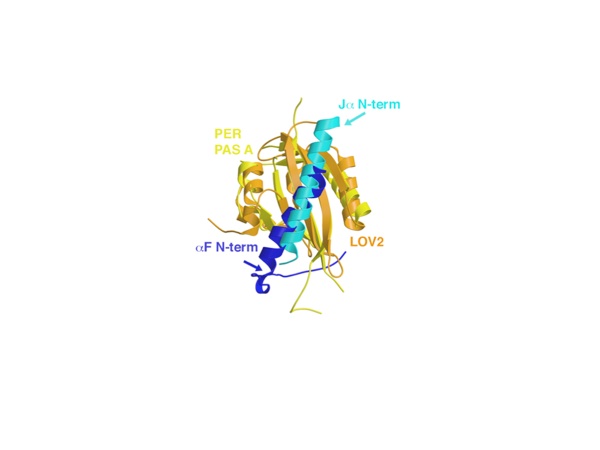
Figure 7. Comparison between αF of PER and the Jα helix of phototropin LOV2. The Jα helix (cyan) of LOV2 PAS (orange, shown here for the Avena sativa protein—2V0U) has similar interactions with the PAS β-sheet as αF (blue) of the PER PAS-A domain (yellow), except that the helices run in opposite directions across the β-sheet. Both helices are known to displace from their respective core PAS domains.
Figure S1. Differences in disulfide bond formation between current PER structure and 1WA9. (A) Compared to 1WA9 the connecting loop after the final strand of the PAS B β-sheet (βE) is held closer to the protein core by a disulfide bridge between C469 on the PAS β-sheet (βD) and C511 of the loop. (B) A closer view shows how the disulfide bond restructures the loop between, βD, and αE.
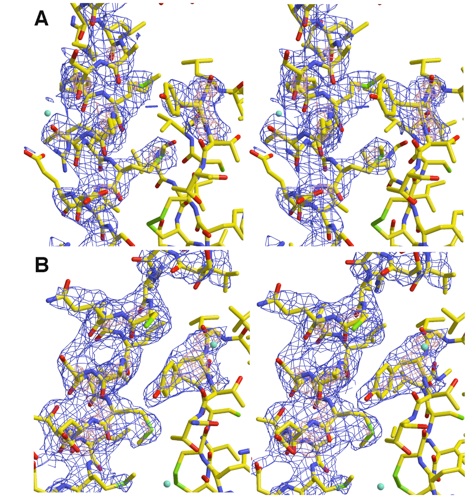
Figure S2. Electron Density for two conformations of αF in PER homodimer. Stereoviews of the two distinct conformations (A and B) of αF in the two PER subunits. 2.85 Å resolution Fo-Fc omit electron density calculated with model phases that do not include the highlighted regions (blue contours at 3σ and red contours at 5σ). Conformation of Tyr253 alters in response to the change in αF angle and register.
Figure S3. Alignment of PER sequences.
Figures from the paper:
Coordinates:
Abstract:
Period (PER) is the major transcription inhibitor in metazoan circadian clocks and lies at the center of several feedback loops that regulate gene expression. Dimerization of Drosophila PER influences nuclear translocation, repressor activity, and behavioral rhythms. The structure of a central, 346-residue PER fragment reveals two associated PAS (Per-Arnt-Sim) domains followed by a protruding α-helical extension (αF). A closed, pseudo-symmetric dimer forms from a cross handshake interaction of the N-terminal PAS domain with αF of the opposing subunit. Strikingly, a shift of αF against the PAS β-sheet generates two alternative subunit interfaces in the dimer. Taken together with a previously reported PER structure in which αF extends, these data indicate that αF unlatches to switch association of PER with itself to its partner Timeless. The variable positions of the αF helix provide snapshots of a helix dissociation mechanism that has relevance to other PAS protein systems. Conservation of PER interaction residues among a family of PAS-AB-containing transcription factors suggests that contacts mediating closed PAS-AB dimers serve a general function.
King, H.A., Hoelz, A., Crane, B.R., Young, M.W. (2011). J. Mol. Biol., 413, 561-572.
Structure of an Enclosed Dimer Formed by the Drosophila
Period Protein






California Institute of Technology
Division of Chemistry & Chemical Engineering
1200 E. California Blvd.
Pasadena, CA 91125-7200
© Copyright Hoelz Laboratory
