Hoelz Lab: Publications
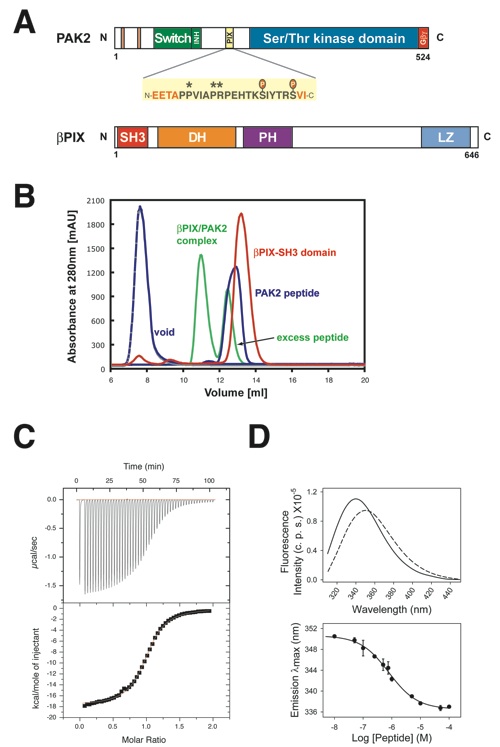
Figure 1. Biochemical analysis of the interaction between the βPIX-SH3 domain and the PAK peptide. (a) Domain structure of PAK2 and βPIX. The PIX binding site is highlighted and the amino acid sequence of the crystallized peptide is shown. Residues shown in black are observed in the electron density map of the complex structure, whereas residues shown in red are missing. Stars above the sequence indicate the conserved PxxxPR motif, and known phosphorylation sites are indicated. (b) Gel filtration on a Superdex 10/300 GL column. The elution profiles for the βPIX-SH3 domain (red) and the PAK peptide (blue) are shown as well as the complex between the two molecules (green). (c) Representative result of isothermal calorimetric titration measurements of the βPIX-SH3 domain with the PAK peptide. Upper panel, raw data of the titration of the SH3 domain (50 μM) with WT PAK peptide (500 μM). Lower panel, dilution corrected integrated heat change and fitted curve base on a single-site binding model. Binding parameters for WT and mutant PAK peptides are presented in Table 2. (d) Upper panel, steady-state fluorescence emission spectra of 1 μM βPIX-SH3 domain in the absence (broken line) and presence (continuous line) of 100 μM WT PAK peptide. Lower panel, titration of βPIX-SH3 domain with increasing amounts of WT PAK peptide. Binding parameters for WT and mutant PAK peptides are presented in Table 2.

Hoelz, A.*, Janz, J.M., Lawrie, S.D., Corwin, B., Lee, A., Sakmar, T.P.*
(2006). J. Mol. Biol. 358, 509-522.
* Corresponding authors
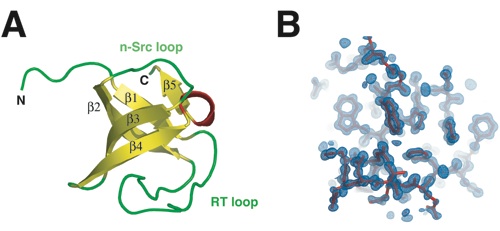
Figure 2. The structure of the βPIX-SH3 domain. (a) Structure of the βPIX-SH3 domain from rat is shown in ribbon representation. Secondary structure elements are labeled. (b) A representative portion of the 2Fo−Fc electron density map, sliced at 4.5 s (light blue), and the SH3 domain model, illustrated in stick representation (red), are shown.
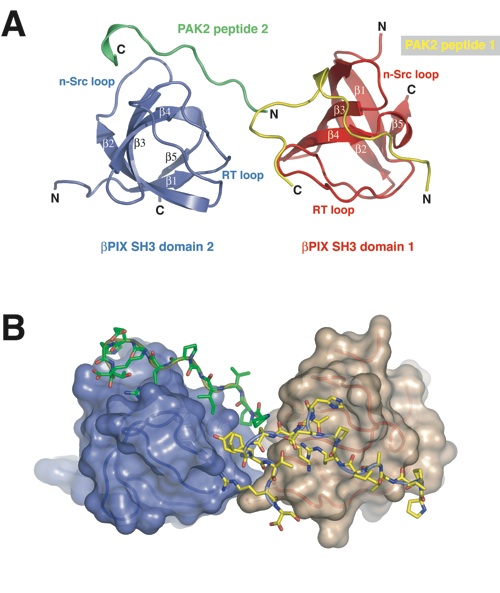
Figure 3. The structure of the βPIX-SH3 domain in complex with a PAK2-derived peptide. (a) Structure of the βPIX-SH3/PAK2 complex shown in ribbon representation as found in the asymmetric unit of the crystal. The two βPIX-SH3 domains are illustrated in red and blue, and the two peptides are illustrated in yellow and green. (b) The surfaces of the βPIX-SH3 domains are shown in van der Waals representation, and the peptides in stick representation. The colors are as in (a).
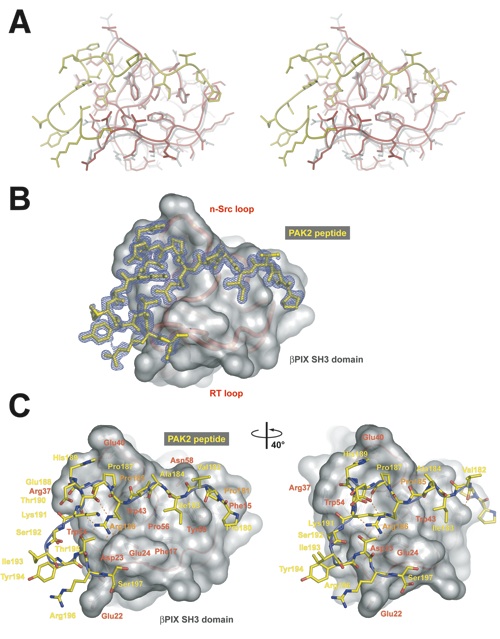
Figure 4. The interface between the βPIX-SH3 domain and the PAK2-derived peptide. (a) Stereo view of the structure of the βPIX-SH3/PAK2 complex (red and yellow) which is superimposed on the structure of the βPIX-SH3 domain (gray). (b) The simulated-annealed 2Fo−Fc electron density map, sliced at 2.5 s, is shown for the PAK peptide (yellow stick representation). The Cα trace of the βPIX-SH3 domain is shown in red below the transparent van der Waals representation of the surface (gray). (c) Close-up view of the entire interface. PAK2 residues are labeled in yellow, and βPIX-SH3 domain residues are labeled in red. Residues in PAK2 that are essential for the interaction are labeled in orange.
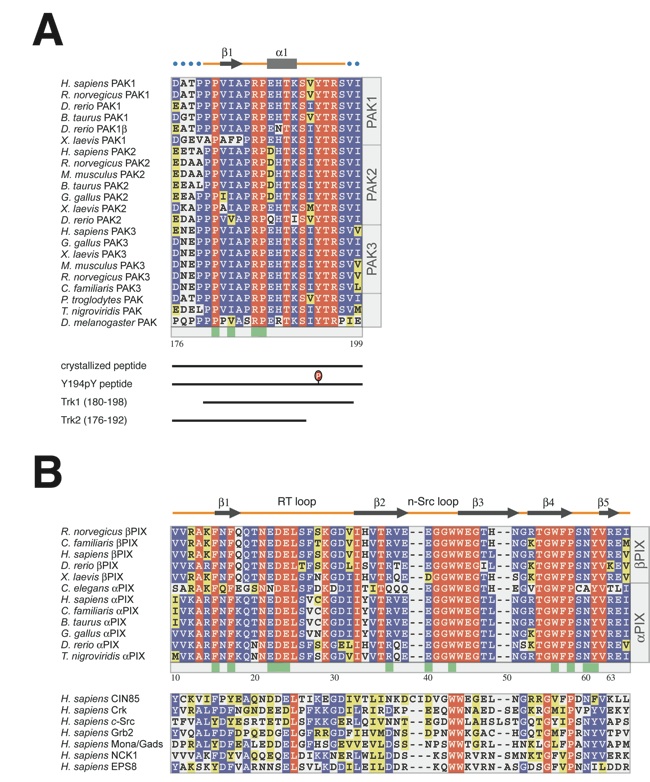
Figure 5. Sequence and structure alignment. Sequence alignment of PIX-binding sequences in PAK kinases (a), PIX-SH3 domains ((b), top panel), and non-PIX family SH3 domains ((b), bottom panel). Sequence numbering corresponds to human PAK2 and rat βPIX. Residues in the alignment are color-coded according to their conservation. Identical residues are shown in red, and conserved residues in blue and yellow. Secondary structure elements are indicated above the sequences, with blue dots indicating disordered residues. The participation of various residues in the formation of the interface between the two molecules is indicated by green boxes below the aligned sequences.
PDB coordinates (link to PDB site) - Space Groups P61, P21212
PDB coordinates - Space Groups P61 (.pdb), P21212 (.pdb)
Structure Factors - Space Groups P61 (.txt), P21212 (.txt)
Figures from the paper:
Coordinates:
Abstract:
The p21-activated kinases (PAKs) are important effector proteins of the small GTPases Cdc42 and Rac and control cytoskeletal rearrangements and cell proliferation. The direct interaction of PAKs with guanine nucleotide exchange factors from the PIX/Cool family, which is responsible for the localization of PAK kinases to focal complexes in the cell, is mediated by a 24-residue peptide segment in PAKs and an N-terminal src homology 3 (SH3) domain in PIX/Cool. The SH3-binding segment of PAK contains the atypical consensus-binding motif PxxxPR, which is required for unusually high affinity binding. In order to understand the structural basis for the high affinity and specificity of the PIX-PAK interaction, we solved crystal structures for the N-terminal SH3 domain of betaPIX and for the complex of the atypical binding segment of PAK2 with the N-terminal SH3 domain of betaPIX at 0.92 A and 1.3A resolution, respectively. The asymmetric unit of the crystal contains two SH3 domains and two peptide ligands. The bound peptide adopts a conformation that allows for intimate contacts with three grooves on the surface of the SH3 domain that lie between the n-Src and RT-loops. Most notably, the arginine residue of the PxxxPR motif forms a salt-bridge and is tightly coordinated by a number of residues in the SH3 domain. This arginine-specific interaction appears to be the key determinant for the high affinity binding of PAK peptides. Furthermore, C-terminal residues of the peptide engage in additional interactions with the surface of the RT-loop, which significantly increases binding specificity. Compared to a recent NMR structure of a similar complex, our crystal structure reveals an alternate binding mode. Finally, we compare our crystal structure with the recently published betaPIX/Cbl-b complex structure, and suggest the existence of a molecular switch.
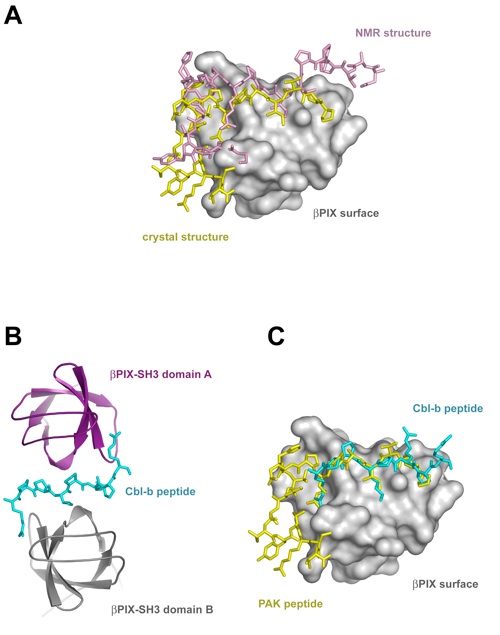
Figure 6. Comparison of the βPIX-SH3/PAK2 structure to other βPIX-SH3 structures. (a) Superposition of the crystal and NMR structures of the βPIX-SH3/PAK complex. Peptides are illustrated in stick representation. The βPIX-SH3 surface is shown in gray, and the peptides are colored according to the legend in (a). (b) Structure of the βPIX-SH3/Cbl-b complex shown in ribbon representation (PDB code 2AK5). The two βPIX-SH3 domains and the Cbl-b peptide are colored according to the legend in (b). (c) Superposition of the βPIX-SH3/PAK2 crystal structure with the βPIX-SH3 B/Cbl-b crystal structure. βPIX-SH3 A has been removed for clarity. Peptides are illustrated in stick representation. The peptides of the crystal structures are colored according to the legend in (c).

Crystal structure of the SH3 domain of βPIX in complex
with a high affinity peptide from PAK2
California Institute of Technology
Division of Chemistry & Chemical Engineering
1200 E. California Blvd.
Pasadena, CA 91125-7200
© Copyright Hoelz Laboratory
