Hoelz Lab: Publications
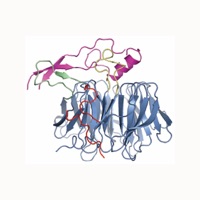
Figure 1. Structural overview of the Rae1Nup98GLEBS complex. (A) Domain organization of human Rae1 and human Nup98. For Rae1, the N-terminal extension (NTE, red) and the seven WD40 repeats (orange) are indicated. For Nup98, the GLEBS motif (magenta), the FG-repeat region (gray), the unstructured region (dark gray), the autoproteolytic domain (pink), and the C-terminal 6 kDa fragment (light gray) that is removed by co-translational proteolysis are indicated. In an alternatively spliced version, the Nup98-96 precursor, the 6 kDa fragment is replaced by Nup96, a protein that is embedded in the symmetric NPC core (Fig. S1). The arrows indicate the sites of autoproteolytic cleavage. (B) Ribbon representation of the Rae1Nup98GLEBS complex. The Nup98 GLEBS motif is indicated in magenta. For Rae1, the beta-propeller domain (blue), the NTE (red), the 5D6A loop (green), and the 7BC loop (yellow) are indicated. A 90° rotated view is shown on the right. (C) Schematic representation of the Rae1Nup98GLEBS structure, colored according to panel B. The blades of the Rae1 beta propeller are labeled from 1 to 7. An asterisk indicates the Velcro-closure beta-strand (17).

Ren, Y., Seo, H.S., Blobel, G., Hoelz, A. (2010). Proc. Natl. Acad. Sci. USA 107, 10406-10411.
Figure 2. Surface properties of Rae1 and the Rae1Nup98GLEBS complex. (A) Surface renditions of Rae1 and Rae1Nup98GLEBS. The Rae1 surface that mediates the association with the Nup98 GLEBS motif is colored in magenta; the remainder is colored in blue. In the Rae1Nup98GLEBS complex, the Rae1 and the Nup98 GLEBS motif surfaces are colored in blue and light magenta, respectively (right panel). As a reference, a ribbon representation of the Rae1Nup98GLEBS complex is shown and corresponds to the orientation of the left panel. The black line in the left panel indicates the location of the side view surface. (B) The surface representations of Rae1 and the Rae1Nup98GLEBS complex are colored according to multispecies sequence alignments (Fig. 4A and Fig. S3). The conservation at each position is mapped onto the surface and shaded in a color gradient from yellow (60 % similarity) to red (100 % identity). (C) Rae1 and Rae1Nup98GLEBS surface renditions, colored according to the electrostatic potential, ranging from red (-10 kBT/e) to blue (+10 kBT/e).
Figure 3. Structural comparison between the Rae1Nup98 and Bub3Mad3 complexes. The superposition of the Rae1 and Bub3 beta-propeller domains is shown in the right panel. Bub3 and Mad3 are colored in gray and pink, respectively. The 5D6A inter-blade connectors and 7BC loops of the Rae1 and Bub3 beta propellers are indicated in different shades of green and yellow, respectively. A 90° rotated view is shown in the lower panel.
Figure 4. The tandem glutamate element of the Nup98 GLEBS motif is essential for the interaction with Rae1. (A) Multispecies sequence alignment of the Nup98 GLEBS motif. The overall sequence conservation at each position is shaded in a color gradient from yellow (40 % similarity) to red (100 % identity) using the Blosum62 weighting algorithm (40). The numbering of the residues and the secondary structure are according to human Nup98. The secondary structure is indicated above the sequence as green arrows (beta-strands), blue rectangles (alpha-helices), gray lines (coil regions), and gray dots (disordered residues). Dots below the sequence indicate residues involved in Rae1 binding (magenta and green). Residues that are shown in panel B are highlighted with magenta dots. Asterisks indicate the positions of the invariant Glu201 and Glu202 of the tandem glutamate element. (B) Details of the interaction between Rae1 and the Nup98 GLEBS motif. The ribbon representation is colored according to Figure 1B. The inset illustrates the position of the tandem glutamate element and is expanded on the right. (C) Analysis of the interaction between Rae1 and GST-Nup98 GLEBS fragments and mutants. The C-terminal arm of the Nup98 GLEBS (GLEBS-C) is necessary and sufficient for Rae1 binding. A double mutant in GLEBS-C, E201K/E202K, abolishes the Rae1-Nup98GLEBS interaction.
Figure 5. Rae1Nup98GLEBS binds RNA in vitro. An electrophoretic mobility shift assay was carried out, with a constant amount of degenerate 10-mer RNA oligonucleotide (2 uM) and increasing concentrations of the Rae1Nup98GLEBS complex, as indicated. The RNA oligonucleotide was visualized with SYBR Gold nucleic acid gel stain. The estimated dissociation constant of the interaction is in the low uM range.
Figure 6. In vivo analysis of the Gle2-Nup116GLEBS interaction. (A) Yeast growth assay performed using nup116delta cells transformed with the indicated mCherry (mCh)-Nup116 constructs. 10-fold serial dilutions were spotted on SD-LEU plates and grown for 2-3 days at the indicated temperatures. (B) In vivo localization of Gle2-GFP and mCh-Nup116 carried out at 30 ºC. In the presence of full-length mCh-Nup116, Gle2-GFP is enriched at the nuclear envelope. The deltaGLEBS and the tandem glutamate element (E154K/E155K) Nup116 mutants result in a dispersed staining of Gle2-GFP throughout the cell with no enrichment at the nuclear envelope. The scale bar represents 5 um.
Figure S1. Domain structure of Nup98 and the Nup98-96 precursor. For Nup98, the GLEBS motif (magenta), the FG-repeat region (gray), the unstructured region (dark gray), the autoproteolytic domain (APD; pink), and the C-terminal 6 kDa fragment (light gray) that is removed by co-translational proteolysis are indicated. In the Nup98-96 precursor, an alternatively spliced version of Nup98, the 6 kDa fragment is replaced by Nup96, a protein that is embedded in the symmetric NPC core (17, 18) (Fig. S1). The arrows indicate the sites of autoproteolytic cleavage.
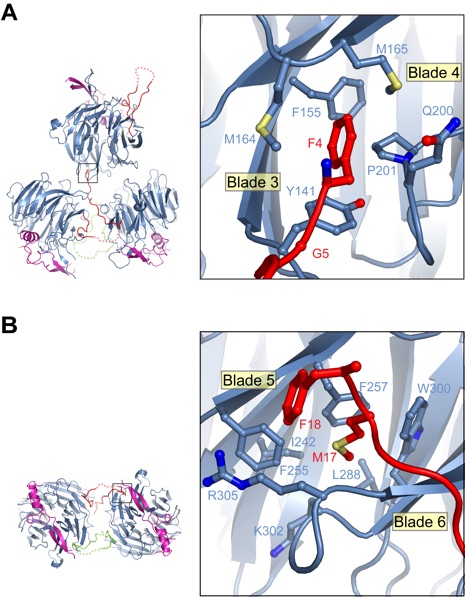
Figure S3. Crystallographic interactions mediated by the NTE of Rae1. (A) Interactions between the four Rae1Nup98GLEBS complexes in the asymmetric unit are illustrated. For clarity, the fourth copy of the complex is omitted. The N-terminal FG-repeat-like sequence (residues 4 and 5) of a Rae1 molecule binds to a hydrophobic pocket of an adjacent Rae1 molecule. A close-up view of the FG-binding pocket is shown in the right inset. (B) Two Rae1Nup98GLEBS complexes interact via a handshake mode mediated by the Rae1 NTE. A 90° rotated view of the bottom two molecules of panel A is shown. The MFG sequence stretch (residues 17-19) of the Rae1 NTE binds to a hydrophobic pocket of a neighboring Rae1 molecule. A close-up view of the interaction is shown in the right inset. No electron density is observed for Gly19 of Rae1.
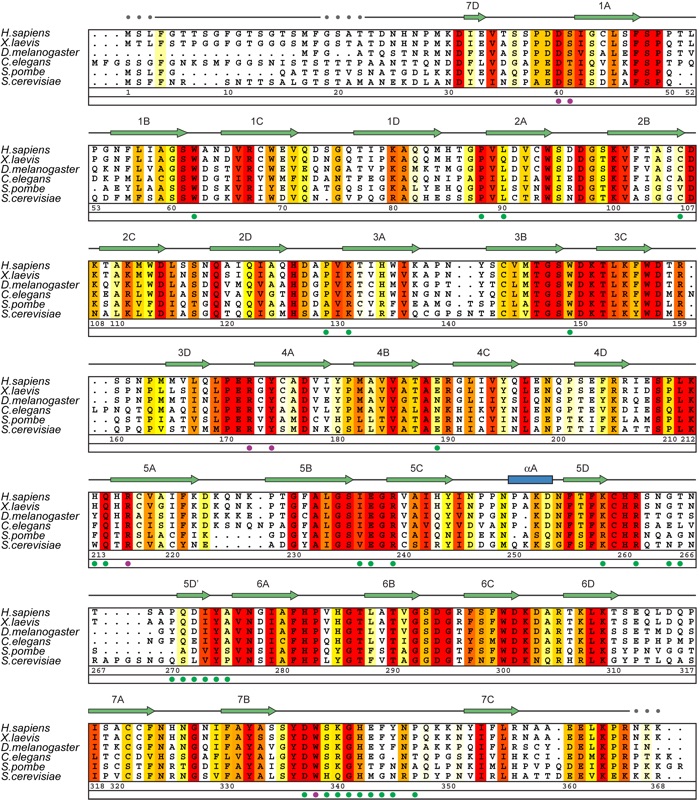
Figure S4. Multispecies sequence alignment of Rae1 homologs. The numbering of the residues and the secondary structure are according to human Rae1. The secondary structure is indicated above the sequence as green arrows (beta-strands), blue rectangles (alpha-helices), gray lines (coil regions), and gray dots (disordered residues). The overall sequence conservation at each position is shaded in a color gradient from yellow (60 % similarity) to red (100 % identity) using the Blosum62 weighting algorithm (19). The Rae1 residues that are involved in the interaction with Nup98 GLEBS motif are indicated with dots below the alignment. The residues that are labeled with magenta dots are shown as sticks in Figure 4B.
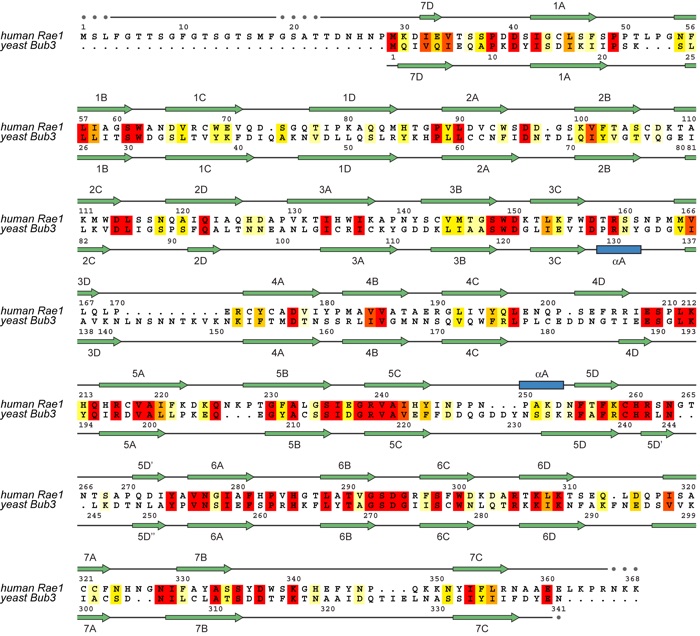
Figure S5. Structure-based alignment of the primary sequence of human Rae1 and yeast Bub3. Corresponding secondary structure elements of the two beta propellers are indicated above (Rae1) and below (Bub3) the aligned sequences.
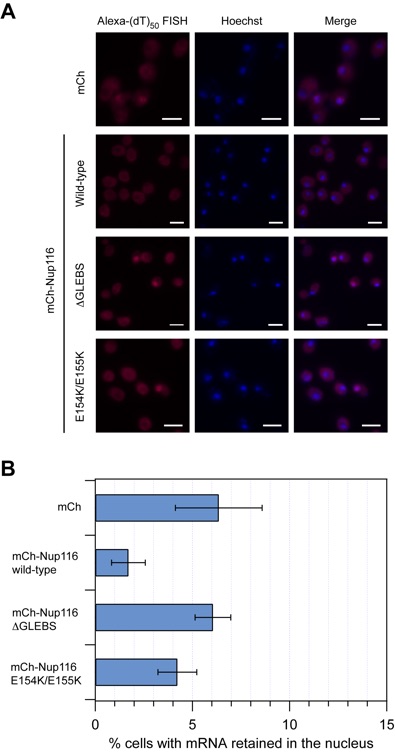
Figure S6. mRNA export assay of Nup116 mutants. (A) Detection of poly(A) mRNA and DNA was carried out using an Alexa-647- labeled 50-mer oligo dT FISH probe and the Hoechst DNA stain, respectively. The merging of the two images is shown on the right. The scale bar represents 5 μm. (B) Quantitation of nuclear poly(A) mRNA retention. The percentages refer to the fraction of cells displaying marked nuclear staining. The error bars correspond to the standard deviation and are derived from four independent images. The P-values of the Nup116 deltaGLEBS and E154K/E155K mutants are less than 0.05, compared to the wild-type Nup116 construct (0.00048 and 0.00936, respectively), indicating that the observed differences between these strains is statistically significant.
Figures from the paper:
Coordinates:
Abstract:
The export of mRNAs is a multistep process, involving the packaging of mRNAs into messenger ribonucleoprotein particles (mRNPs), their transport through nuclear pore complexes, and mRNP remodeling events prior to translation. Ribonucleic acid export 1 (Rae1) and Nup98 are evolutionarily conserved mRNA export factors that are targeted by the vesicular stomatitis virus matrix protein to inhibit host cell nuclear export. Here, we present the crystal structure of human Rae1 in complex with the Gle2-binding sequence (GLEBS) of Nup98 at 1.65 angstroms resolution. Rae1 forms a seven-bladed beta-propeller with several extensive surface loops. The Nup98 GLEBS motif forms an approximately 50-angstroms-long hairpin that binds with its C-terminal arm to an essentially invariant hydrophobic surface that extends over the entire top face of the Rae1 beta-propeller. The C-terminal arm of the GLEBS hairpin is necessary and sufficient for Rae1 binding, and we identify a tandem glutamate element in this arm as critical for complex formation. The Rae1*Nup98(GLEBS) surface features an additional conserved patch with a positive electrostatic potential, and we demonstrate that the complex possesses single-stranded RNA-binding capability. Together, these data suggest that the Rae1*Nup98 complex directly binds to the mRNP at several stages of the mRNA export pathway.
Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1.

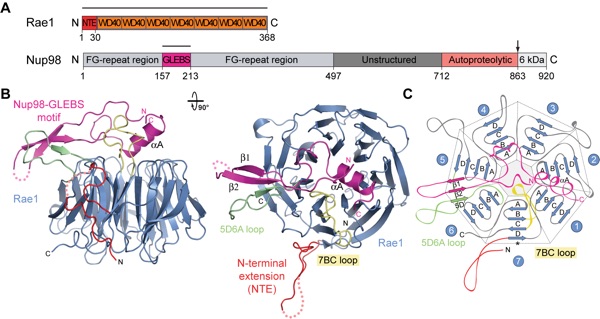
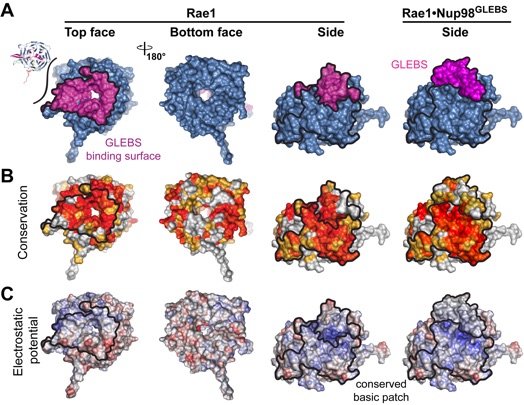
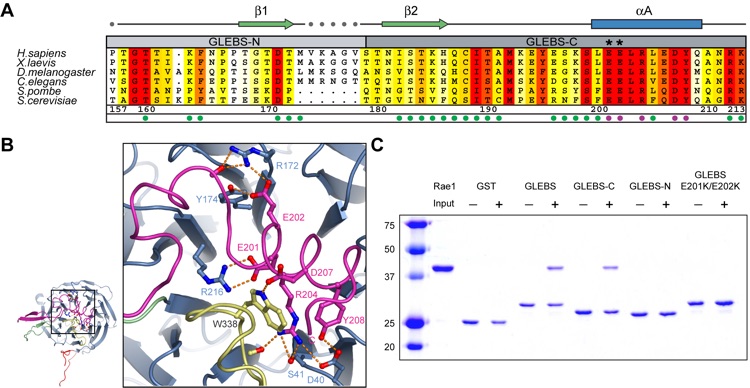
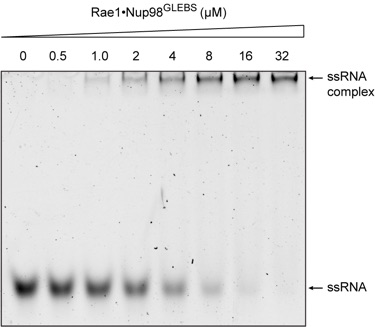
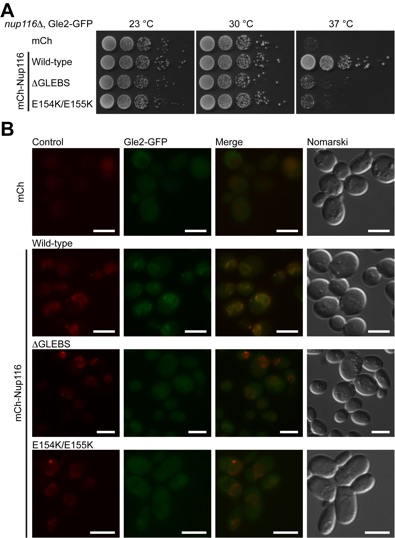
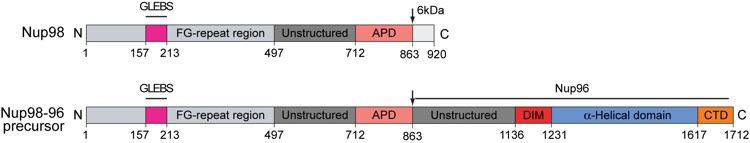
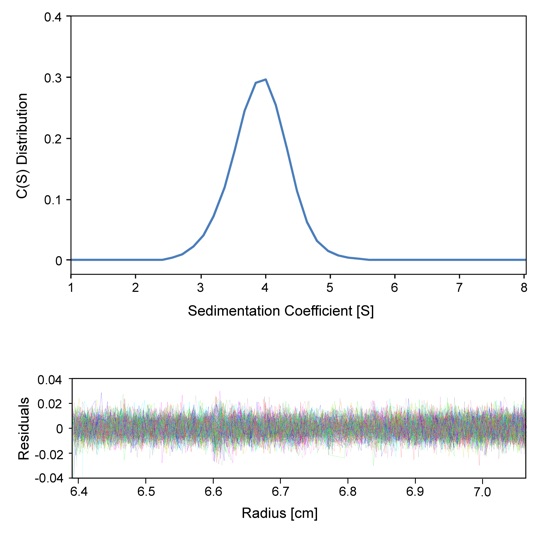
Figure S2. Sedimentation velocity analysis of the Rae1Nup98GLEBS complex. The sedimentation coefficient distribution shows a single species (top panel). The determined molecular mass is 45.4±7.5 kDa, corresponding to a monomeric species of the Rae1Nup98GLEBS heterodimer (calculated 48.4 kDa) in solution. The residuals for the fitting are shown at the bottom panel.
California Institute of Technology
Division of Chemistry & Chemical Engineering
1200 E. California Blvd.
Pasadena, CA 91125-7200
© Copyright Hoelz Laboratory
