Hoelz Lab: Publications
Figure 1. Structure of S. cerevisiae Rtt109ΔL. (A) Domain structure. Blue, protein acetyltransferase (PAT) domain; yellow, activation domain; red, PAT-activation domain connectors; gray, unstructured C-terminal region (U). The bar above the domain structure marks the crystallized fragment. (B) Structure of Rtt109ΔL in ribbon representation, colored as in panel A. A 90°-rotated view is shown on the right.

Stavropoulos, P., Nagy, V., Blobel G., Hoelz A.
(2008). Proc. Natl. Acad. Sci. USA 105, 12236-12241.
Figure 2. Interface between PAT and activation domains. Ribbon representation of the interface between the PAT and the activation domains colored according to Fig. 1A. The activation domain is tightly bound to the PAT domain. The acetylated Lysine-290 of the activation domain is located in the center of the interface between the two domains and is inserted into a hydrophobic cavity of the PAT domain. The active site cavity is located ~10 Å away from the Ac-K290 binding site.
Figure 3. Surface properties of Rtt109ΔL. (A) Surface representation colored according to the participation of the various domains as in Fig. 1B. (B) Surface representation colored according to a multi-species sequence alignment (Fig. S1). The conservation at each position is mapped onto the surface and is shaded in a color gradient from light yellow (40 % similarity) to dark red (100 % identity). (C) Surface representation colored according to the electrostatic potential. The electrostatic potential is plotted onto the surface and colored in a gradient from red (-10kBT/e) to blue (+10kBT/e). The orientation of all surface representations is identical.
Figure 4. Structural comparison of Rtt109ΔL to p300/CBP. (A) Superposition of the structure of Rtt109ΔL (left, colored as in Fig. 1A) in complex with Acetyl-CoA (orange) with p300/CBP (middle, PAT domain in grey and the activation domain in green) in complex with the bi-substrate inhibitor Lysine-CoA (purple). The inset illustrates the position of the active site and is expanded in panel B. (B) Superposition of the active sites of Rtt109 (left) with p300/CBP (middle). Critical active site residues are shown in stick representation, and the Cα-traces are shown in coil representation, according to the coloring scheme in panel A. The orientation of all active sites is identical. The PDB code of the p300/CBP structure is 3BIY (21).
Figure 5. Model for the activation mechanism of Rtt109. The activation of the S. cerevisiae protein acetyltransferase Rtt109 depends on the auto-acetylation of Lysine-290. We propose that Rtt109 exits in two states, an inactive, auto-inhibited state and an activated state. In the inactive state, the activation domain is loosely tethered to the PAT domain. Upon auto-acetylation, the PAT domain engages the acetylated Lysine-290, resulting in the tight interaction between the activation and PAT domains. The catalytic activity of Rtt109 may be further enhanced by the sequestration of the Vps75 interaction loop, removing it from the active site groove.
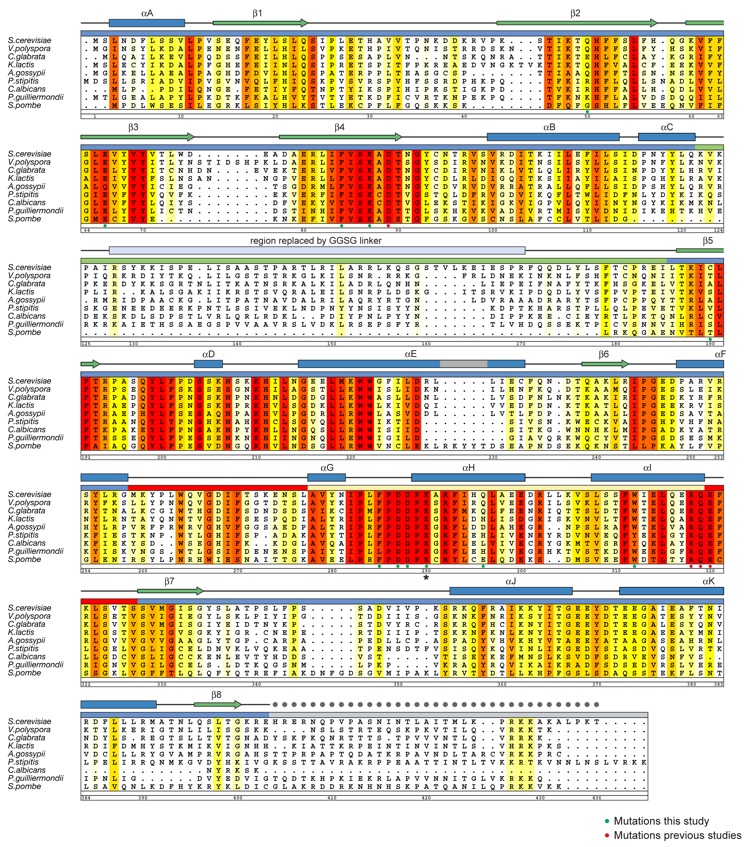
Figure S1. Multi-Species Sequence Alignment of Rtt109 Homologs. The numbering below the alignment is relative to S. cerevisiae Rtt109. The overall sequence conservation at each position is shaded in a color gradient from yellow (40 % similarity) to dark red (100 % identity), using the Blosum62 weighting algorithm. The secondary structure is indicated above the sequence as green arrows (β-strands), blue rectangles (α-helices), gray lines (coil regions), and gray dots (disordered residues). Residues that are important for enzymatic activity (green dots, this study; red dots, previous studies) and the acetylated K290 (star) are indicated below the aligned sequences.
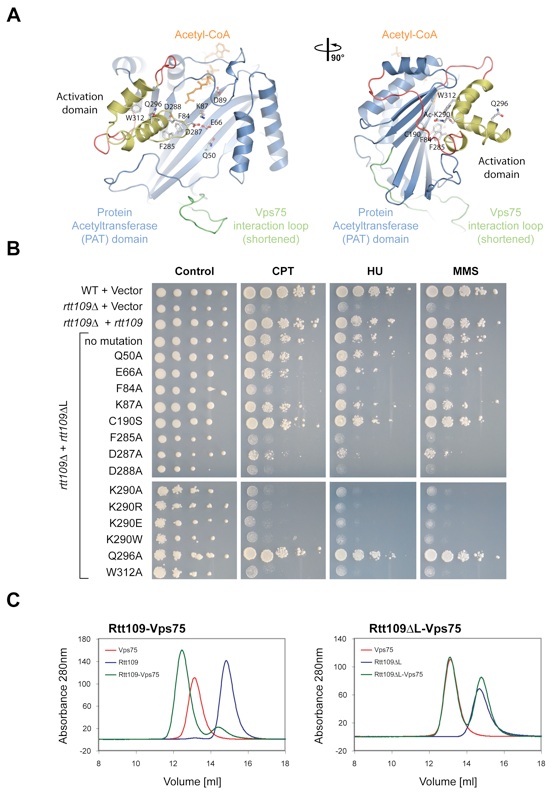
Figure S2. Mutational analysis of Rtt109. (A) Ribbon representation of Rtt109, colored according to Fig. 1A. Mutated residues are illustrated in ball-and-stick representation. A 90°-rotated view is shown on the right. (B) In vivo analysis of Rtt109 mutants in a DNA damage hypersensitivity assay. Serial dilutions (10-fold) of wild type or rtt109 cells transformed with wild type or the indicated mutants were spotted on SD-URA plates alone or on SD-URA plates containing either camptothecin (CPT) at 1 g/ml, hydroxyurea (HU) at 7.6 mg/ml, or methyl methanesulfonate (MMS) at 65 g/ml. (C) Biochemical analysis of the interaction between Rtt109 and Vps75. Gel filtration profiles of full-length Rtt109, Vps75, and their complex (left panel) and Rtt109ΔL, Vps75, and their co-injection (right panel). The Rtt109-Vps75 interaction is dependent on the Vps75 interaction loop. In the Rtt109ΔL structure, the Vps75 interaction loop is genetically shortened and does not contain the Vps75 interaction region.
Figures from the paper:
Coordinates:
Abstract:
Rtt109 is a protein acetyltransferase (PAT) that is responsible for the acetylation of lysine-56 of histone 3 (H3K56) in yeast. H3K56 acetylation has been implicated in the weakening of the interaction between the histone core and the surrounding DNA in the nucleosomal particle. Rtt109, in cooperation with various histone chaperones, promotes genomic stability and is required for resistance to DNA damaging agents. Here, we present the crystal structure of Rtt109 in complex with acetyl-CoA at a 2.0-A resolution. Rtt109 consists of a core PAT domain, which binds the acetyl-CoA cofactor. A second domain, the activation domain, is tightly associated with the PAT domain. Autoacetylation of lysine-290 within the activation domain is required for stabilizing the interaction between the two domains and is essential for catalysis. Biochemical analysis demonstrates the requirement of a loop within the PAT domain for the binding of the histone chaperone Vps75, and mutational analysis identifies key residues for catalysis. We propose a model in which the autoacetylation of Rtt109 is crucial for the regulation of its catalytic activity.
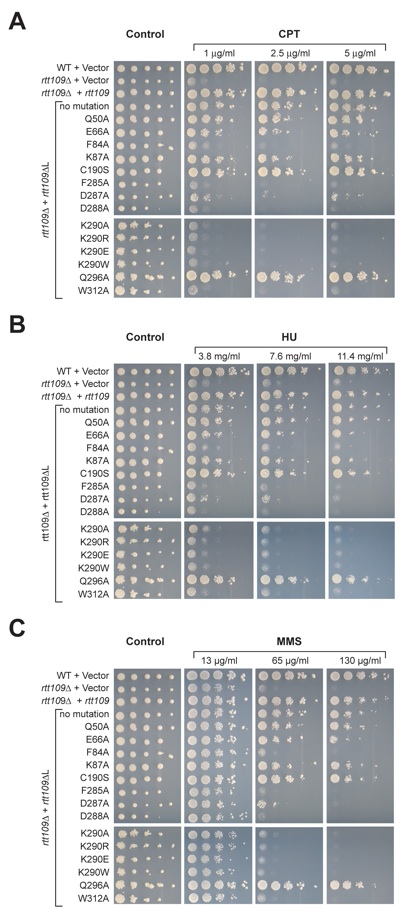
Figure S3. Mutational analysis of Rtt109 in a DNA damage hypersensitivity assay. Serial dilutions (10-fold) of wild type or rtt109 cells transformed with wild type or the indicated mutants were spotted on SD-URA plates alone or on SD-URA plates containing either (A) camptothecin (CPT) at a concentration of 1-5 g/ml, (B) hydroxyurea (HU) at a concentration of 3.8-11.4 mg/ml, or (C) methyl methanesulfonate (MMS) at a concentration of 13-100 g/ml.







Molecular basis for the autoregulation of the protein acetyl transferase Rtt109.
California Institute of Technology
Division of Chemistry & Chemical Engineering
1200 E. California Blvd.
Pasadena, CA 91125-7200
© Copyright Hoelz Laboratory
