Hoelz Lab: Publications
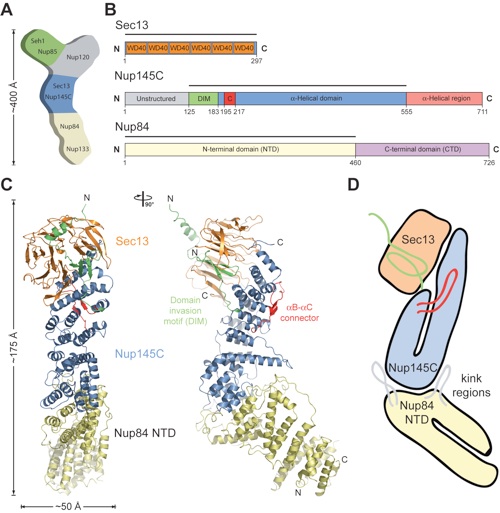
Figure 1. Structure of the S. cerevisiae Sec13•Nup145C•Nup84 NTD complex. (A) Schematic representation of the heptameric complex and the approximate localization of its seven nups (21). (B) Domain structures of Sec13, Nup145C, and Nup84. For Sec13, the six WD40 repeats (orange) are indicated. For Nup145C, the unstructured N-terminal region (gray), the domain invasion motif (DIM) (green), the αB-αC connector (C) (red), the α-helical domain (blue), and the C-terminal α-helical region (pink) are indicated. For Nup84, the N-terminal domain (NTD) and C-terminal domain (CTD) are indicated. The residue numbering is shown below and the bars above the domain structures mark the crystallized fragments of the three proteins. (C) Structure of Sec13•Nup145C•Nup84 NTD in ribbon representation, colored as in panel B. A 90°-rotated view is shown on the right. (D) Schematic representation of the Sec13•Nup145C•Nup84 NTD heterotrimer.

Nagy, V., Hsia, K.C., Debler, E.W., Kampmann, M., Davenport, A.M., Blobel, G., Hoelz, A.
(2009). Proc. Natl. Acad. Sci. USA 106, 17693-17698.
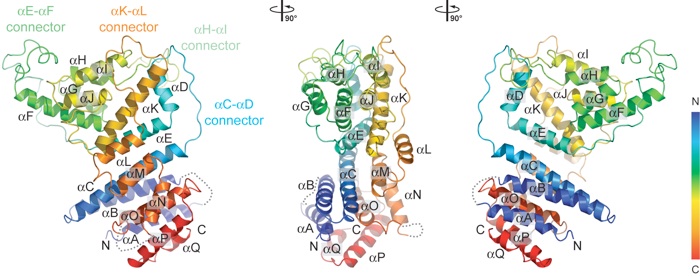
Figure 2. The Nup84 α-helical domain. (A) Ribbon representation of the Nup84 NTD is shown in rainbow colors along the polypeptide chain from the N- to the C-terminus. The four loops that participate in the Nup145C•Nup84 interaction are indicated.
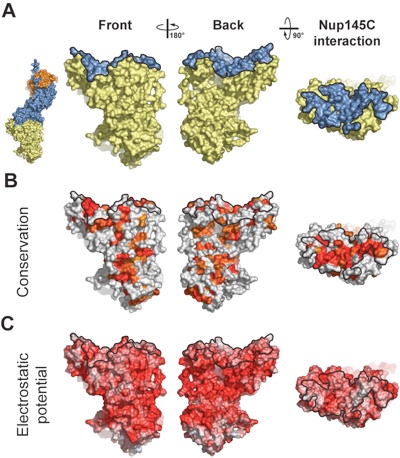
Figure 3. Surface properties of the Nup84 NTD. The surface orientations are identical in all columns. A black line encircles the Nup145C interaction surface. (A) Surface rendition of the Nup84 NTD. The Nup145C contact surface is colored in blue, while the remaining surface is colored in yellow. As a reference, a surface rendition of the heterotrimer is shown to the left, colored according to Fig. 1C. (B) Surface representation colored according to a multi-species sequence alignment, ranging from 60% similarity (white) to 100% identity (red) (Fig. S4). (C) Surface rendition colored according to the electrostatic potential, ranging from -10 kBT/e (red) to +10 kBT/e (blue). Note the two conserved hydrophobic patches located towards the periphery of the extended Nup145C interacting surface.
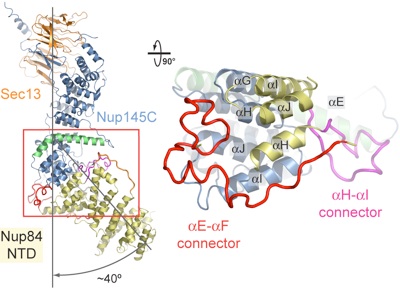
Figure 4. Interaction of the Nup84 NTD with the Nup145C solenoid domain. The Sec13•Nup145C•Nup84 NTD heterotrimer is shown in ribbon representation, colored according to Fig. 1C. The kink regions of the two solenoids interact in a head-to-head fashion. The Nup84 NTD protrudes with a ≈40° angle from the Nup145C U-shaped solenoid. The inset marks the Nup145C•Nup84 interface that is illustrated in detail on the right. For clarity, the interface shown on the right is rotated by 90°. For Nup145C, the solenoid subdomain (blue) and helix αE (green) are indicated. For Nup84, the interface helices (yellow), as well as the long αE-αF (red) and αH-αI (magenta) connectors that mediate the interaction with Nup145C are indicated.
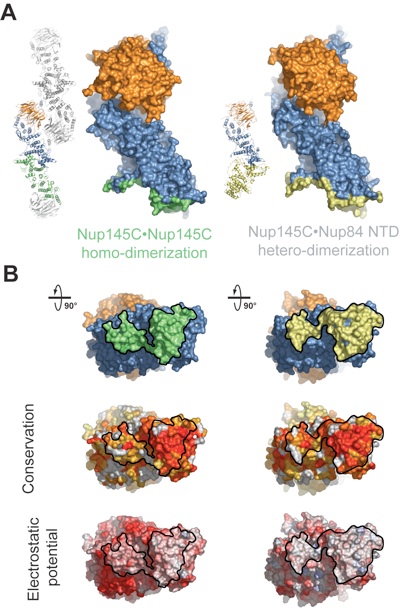
Figure 5. Binding promiscuity of Nup145C. (A) Surface rendition of the Sec13•Nup145C nucleoporin pair derived from the Sec13•Nup145C hetero-octamer (Nup145C•Nup145C homo-dimerization) and the Sec13•Nup145C•Nup84 NTD heterotrimer (Nup145C•Nup84 NTD hetero-dimerization). The Nup145C homo-dimerization and hetero-dimerization surfaces are colored in green and yellow, respectively. The Sec13 and the remaining Nup145C surfaces are colored in orange and blue, respectively. (B) 90°-rotated views of the Sec13•Nup145C pair colored according to panel A (top), to a multi-species sequence alignment, ranging from 60% similarity (white) to 100% identity (red) (26) (middle), and to the electrostatic potential, from -10 kBT/e (red) to +10 kBT/e (blue). The orientation of all surface representations is identical in each column. As a reference, black lines encircle the Nup145C homo-dimerization and Nup84 interaction surfaces.
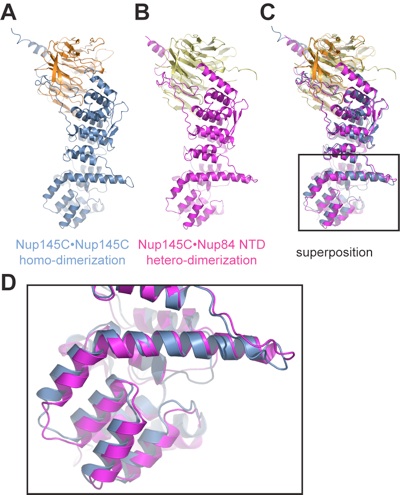
Figure 6. Structural comparison of Nup145C•Nup145C homodimers and Nup145•Nup84 hetero-dimers. (A) Conformation of Sec13•Nup145 in the Sec13•Nup145C hetero-octamer. (B) Conformation of Sec13•Nup145 in the Sec13•Nup145C•Nup84 NTD hetero-trimer. (C) Superposition of the Sec13•Nup145 conformations in panels A and B. The structural alignment is based on Sec13 and the upper arms of the Nup145C solenoid that form a relatively rigid unit. (D) The inset shows the solenoid subdomain (helices αF-J) and the characteristic bent helix αE that undergo conformational changes upon Nup84 NTD binding.
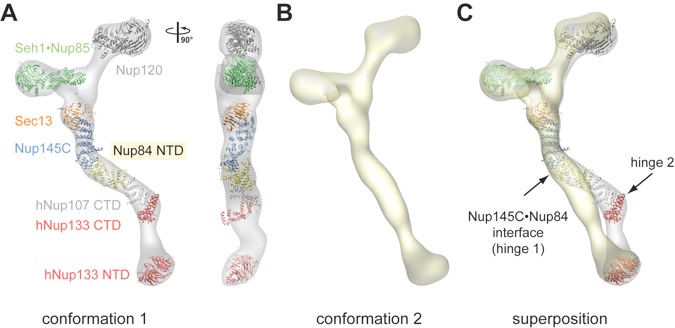
Figure 7. Protein arrangement within the heptameric complex. (A) Docking of crystal structures into the EM envelope of the heptameric Nup84 complex. A 90°-rotated view is shown on the right. The ≈40° angle by which the Nup84 NTD protrudes from the Sec13•Nup145 nucleoporin pair nicely follows one of the two kink regions of the heptamer stem. (B) EM envelope of the second reconstructed conformation of the heptamer in which the two hinge regions are completely extended, forming an almost entirely straight stem. (C) Superposition of the two determined heptamer conformations. The kink region at the Nup145C•Nup84 interface is indicated and was used for the structural alignment, revealing that this interface forms a hinge in the heptamer stem.
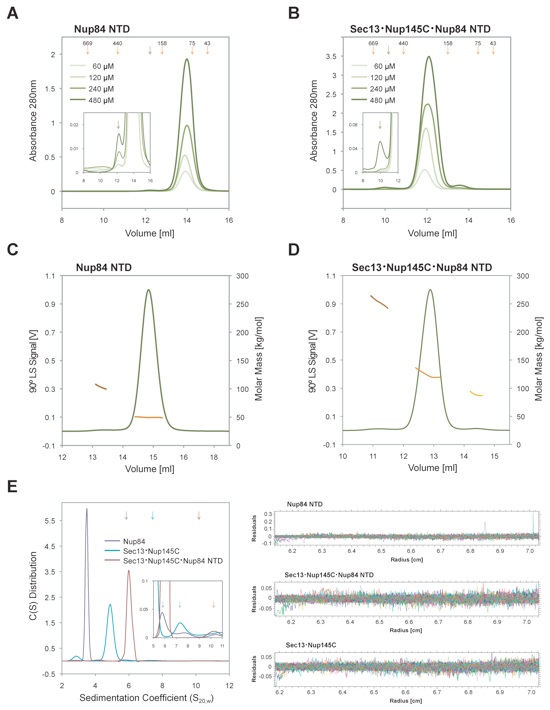
Figure S2. Dynamic behavior of Sec13•Nup145C and Nup84 NTD in solution. Size-exclusion chromatography analysis of (A) the Nup84 NTD and (B) the Sec13•Nup145C•Nup84 NTD complex. All proteins were injected at the indicated concentrations. The elution volumes of molecular weight standards (orange arrows) and the dimer peak positions (green arrows) are illustrated. The inset displays the magnifications of the dimer peaks. Determination of molecular masses by multi-angle light scattering of (C) the Nup84 NTD and (D) the Sec13•Nup145C•Nup84 NTD complex. The 90° light scattering signal is plotted against the elution volume and is overlaid with the determined molecular masses for the selected peaks. The small peak eluting after the principal peak in panels B and D corresponds to excess Sec13•Nup145C. Compared to panels A and B, the peak positions in the light scattering experiments are shifted due to the larger delay volume. (E) Sedimentation velocity analysis of the interaction between Sec13•Nup145C and Nup84 NTD. Sedimentation coefficient distributions of the Nup84 NTD (purple), of the Sec13•Nup145C nucleoporin pair (blue), and the Sec13•Nup145C•Nup84 NTD complex (magenta). Due to the small fraction of the dimeric species in solution combined with the low protein concentration dictated by analytical ultracentrifugation, the dimeric species were only barely detectable. The sedimentation positions of the dimeric species are indicated by arrows in the same color scheme. The residuals of the fits for the three experiments are shown on the right. The molecular masses determined by the various techniques are summarized in Table S2.
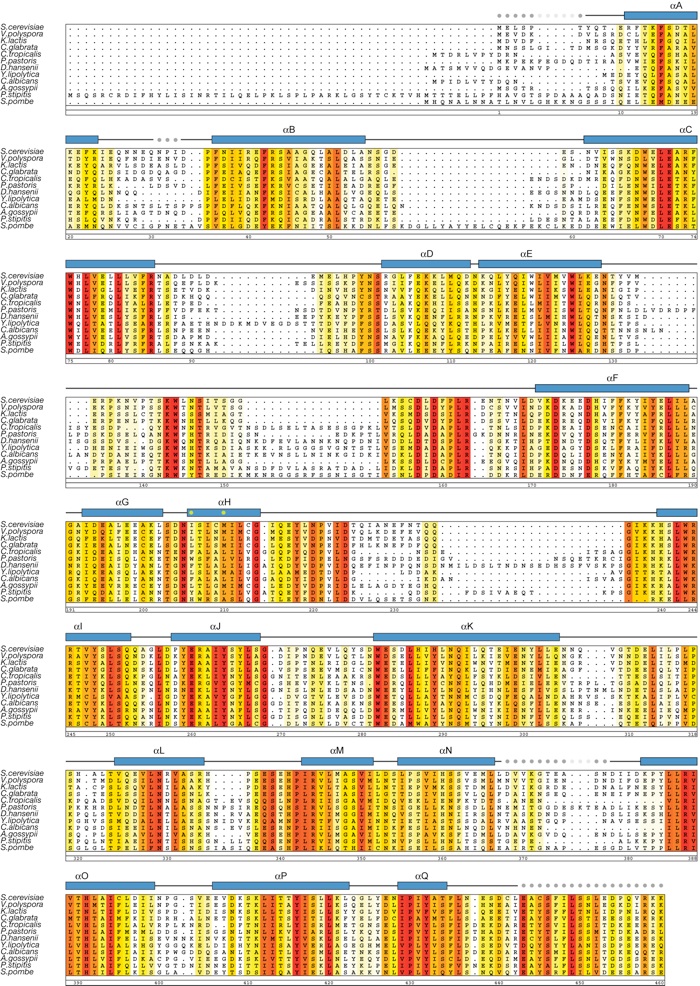
Figure S3. Multi-species sequence alignment of Nup84 homologs. The numbering below the alignment is relative to S. cerevisiae Nup84. The overall sequence conservation at each position is shaded in a color gradient from yellow (60 % similarity) to red (100 % identity) using the Blosum62 weighting algorithm. The secondary structure is indicated above the sequence as blue boxes (α helices), gray lines (coil regions), and gray dots (disordered residues). The positions of the two residues (Ile206 and Met210, located in helix αH) that abolish complex formation with Nup145C when mutated to Asp are indicated by green dots.
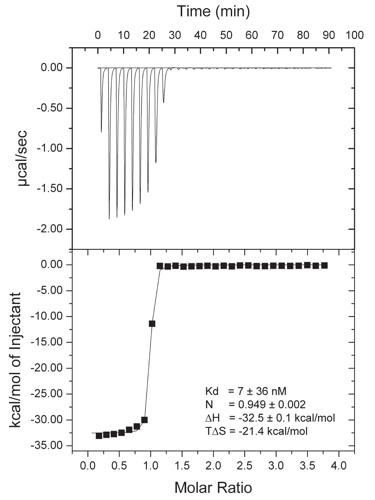
Figure S4. Isothermal titration calorimetry (ITC) analysis of the Sec13•Nup145C•Nup84 NTD complex. The Nup84 NTD was titrated against the Sec13•Nup145C complex. The dissociation constant (Kd), the binding enthalpy (ΔH), and the entropy (TΔS) were derived by curve fitting using the single-site model.
Figures from the paper:
Coordinates:
Abstract:
The heptameric Nup84 complex constitutes an evolutionarily conserved building block of the nuclear pore complex. Here, we present the crystal structure of the heterotrimeric Sec13 x Nup145C x Nup84 complex, the centerpiece of the heptamer, at 3.2-A resolution. Nup84 forms a U-shaped alpha-helical solenoid domain, topologically similar to two other members of the heptamer, Nup145C and Nup85. The interaction between Nup84 and Nup145C is mediated via a hydrophobic interface located in the kink regions of the two solenoids that is reinforced by additional interactions of two long Nup84 loops. The Nup84 binding site partially overlaps with the homo-dimerization interface of Nup145C, suggesting competing binding events. Fitting of the elongated Z-shaped heterotrimer into electron microscopy (EM) envelopes of the heptamer indicates that structural changes occur at the Nup145C x Nup84 interface. Docking the crystal structures of all heptamer components into the EM envelope constitutes a major advance toward the completion of the structural characterization of the Nup84 complex.
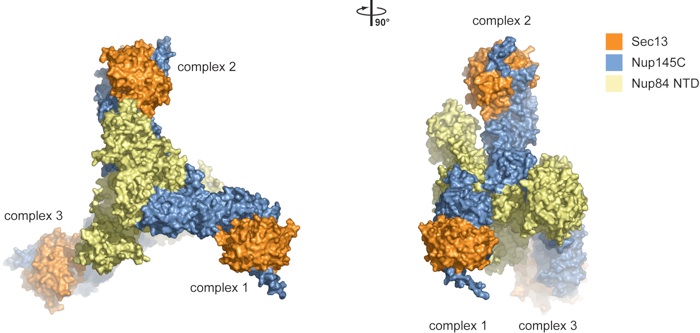
Figure S1. Crystallographic analysis of the Sec13•Nup145C•Nup84 NTD crystals. The asymmetric unit harbors three copies of the Sec13•Nup145C•Nup84 NTD heterotrimer that interact with each other in a head-to-tail fashion and are related by a 32-screw axis. A 90°-rotated view is shown on the right.

Structure of a trimeric nucleoporin complex reveals alternate oligomerization states.
California Institute of Technology
Division of Chemistry & Chemical Engineering
1200 E. California Blvd.
Pasadena, CA 91125-7200
© Copyright Hoelz Laboratory
